At Lexogen we are designing and producing RNA sequencing library preparation kits for use on Illumina sequencing instruments. How RNA-Seq libraries can be generated is described in Chapter 1, our Introduction to RNA Sequencing. In the following chapter we will focus on the sequencing process itself and its underlying principles. In order to interpret library quality parameters, it is helpful to know how the sequencing process known as “Sequencing by Synthesis” functions.
1. Preparing the Library for Sequencing
After the libraries are generated, amplified by PCR, and passed quality control, they are prepared for sequencing. During this process, the concentration is adjusted to the requirements of the sequencer and the double-stranded libraries are denatured to single strands as only single strands can be bound onto the flow cell. Ready-to-sequence libraries contain specific Illumina adapter sequences, termed P5 and P7, at their 5’ and 3’ end (Fig. 1).
These adapter sequences serve two functions:
➊ The “outer” region (shown in black and orange) is required for binding to complementary sequences on the surface of the Illumina flow cell. Thereby, the individual library single strands are captured to be sequenced.
➋ The “inner” region (shown in green and blue) serves as binding site for the sequencing primer which is used to read out the insert sequence during the actual sequencing process.
Figure 1 | Structure of a sequencing-ready Illumina-compatible library. The insert sequence (gray) is flanked by two sequencing adapters. The P5 adapter contains a flow cell binding region (black). This sequence can also coincide with the binding site for the Index 2 sequencing primer for the optional i5 index (Index 2, purple). * Depending on the sequencer the index 2 sequencing primer binding site can be located in the inner or outer region of the adapter. The P5 adapter also contains the Read 1 sequencing primer binding site (green). The P7 adapter contains a flow cell binding region (orange), the i7 index sequence (Index 1, yellow) and the Read 2 / Index 1 sequencing primer binding sites (blue).
2. Cluster Generation
In the first step, the single-stranded sequencing-ready libraries are loaded onto the flow cell. The complementary oligonucleotides on the flow cell surface act as an anchor to capture the libraries by binding to the outer region of the Illumina adapter sequence. Once attached, clusters can be generated, and libraries are sequenced by synthesis.
Cluster generation begins by bridge amplification. During this process, the complementary strand is generated by elongating the oligo attached to the flow cell. Thereby, a copy of the molecule to be sequenced is now covalently attached to the flow cell. The original molecule is washed away, and the strand bends over (like a “bridge”) to attach to the next flow cell oligo. This second oligo is complementary to the other sequencing adapter, and thus upon elongation the reverse strand is generated. After forward and reverse strands are generated and both are stably attached to the flow cell, clusters are generated by clonal amplification, i.e., the process is repeated over and over until the required level of amplification is reached. Essentially, enough material needs to be generated by clonal amplification so that the signals generated in the sequencing process become detectable.
3. Sequencing by Synthesis
First, reverse strands are removed before sequencing starts. This ensures that only the forward strands are sequenced and the read out is homogenous and not overlayed with a second sequence which would otherwise render the detected sequence unusable.
A polymerase then “sequences” the insert of the library by adding nucleotides to the complementary strand that are fluorescently labelled. Depending on the chemistry used in the respective machine, either all four nucleotides are labelled differently, or only a subset of the nucleotides contain a label. The label also acts as a terminator, so that the reaction is stopped after the incorporation of one nucleotide. Figure 2 illustrates the process with four fluorescently labeled nucleotides (Courtesy of Illumina, Inc.). After each cycle, the emission of the fluorophore is detected for each cluster using an optical system and thus the identity of the incorporated base is determined. The termination block and the nucleotides of the previous cycle are removed, and the process is repeated until the desired read length is reached. After completion of the sequencing run, the optical signals are translated into the actual sequence, a process termed base calling (Fig. 2).
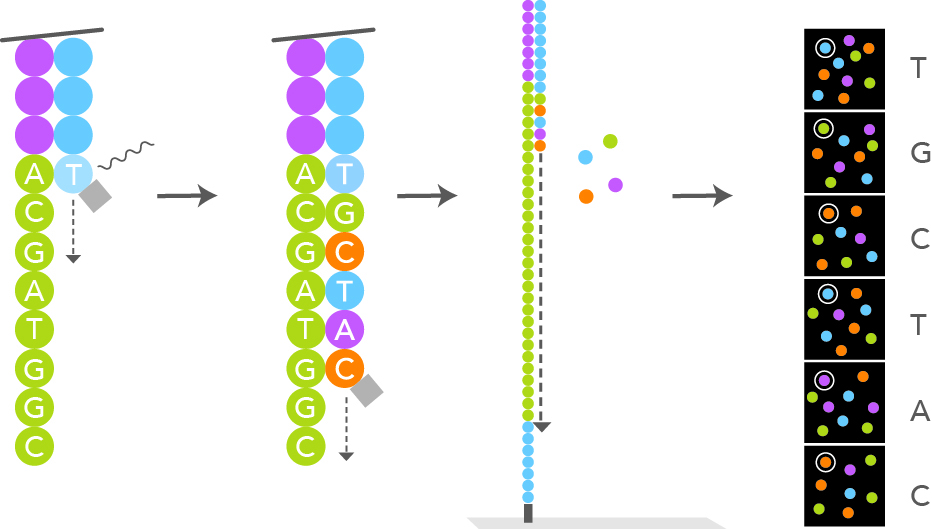
Image Courtesy of Illumina, Inc.
Figure 2 | Sequencing by Synthesis. Sequencing by synthesis technology uses fluorescently labeled A, C, T, and G nucleotides. For each sequencing cycle, a single labeled nucleotide is added to the growing nucleic acid chain. The nucleotide label serves as a terminator for polymerization and after each incorporation the fluorescent dye is imaged. Removal of the dye prior to the subsequent cycle allows incorporation of the next nucleotide. The imaged fluorescent signals are then translated into the nucleic acid sequence. For more information, visit Illumina website.
Following sequencing of the forward strand with the Read 1 primer, the newly created strand is removed. The first index is then read out following the same principle, Index Read 2 is optional. Finally, the reverse strand is read out using the Read 2 primer. To learn more about Sequencing by Synthesis, we recommend the educational resources provided by Illumina.
4. NGS-inherent Sampling Variance
One key facet of sequencing lies in the relationship between the amplification and QC of the libraries, and the sequencing run itself. One way to look at this is by working backwards, starting with the needs of the sequencer – more specifically, the flow cell. For sequencing itself to occur, the lane mix of prepared libraries is strongly diluted following library preparation.
For example, a common scenario would involve diluting a lane mix down to 2 nM, followed by a second dilution by a factor of around 1,000 to a concentration of 2 pM which is used to load on the sequencer. Yet another “dilution” occurs on the flow cell itself, where only a subset of the molecules is actually captured on the flow cell.
So, what does this mean for your library preps? It means that the final “yield” of a library prep method is only really important for one thing, and that is quality control (QC). The last step of library generation is PCR, in which the adapter sequences are completed, indices are introduced, and the library is amplified. Since the ultimate output of a library prep method is subsequently diluted to the diminutive amounts required by a flow cell, it is only necessary to generate enough material to be analyzed by quality control instrumentation.
Library quality control quantifies two primary aspects of a library output: concentration and size distribution of each final library. When it comes to library yield, the amount of material needed to reach the detection limit of the QC instruments is much higher than the requirements of a flow cell as described above.
There is an interesting issue that arises when a library is first amplified during PCR and subsequently diluted for sequencing. This “random drawing” effect is the ultimate determinant of which molecules get sequenced from each sample (Fig. 3). There are a few implications of this effect which we will now review.
5. Why less is more
Graphic drawing based on Illumina’s NextSeq 500
Figure 3 | Only a fraction of the molecules in an NGS-library will be sequenced.
When one library is sequenced twice on separate runs, it is expected that these runs would yield slightly different results in downstream data analysis. As such, the inclusion of controls in each sequencing run is imperative. The sequencing process itself adds technical noise and has an inherent statistical limit. That is, sequencing data has a ceiling that is reached prematurely, as unavoidable noise prevents an inherently perfect sequencing run.
In today’s sequencers, data is stored as a FASTQ file. This data is comprised of the collective reads from a sequencing run as well as other statistical measures of the quality of the sequencing run, i.e., the sequencer’s own confidence assessment. The factors that impact this quality assessment are a result of the imperfections associated with the sequencing process itself. There are some unavoidable adverse effects which occur regularly, though in small numbers, in any given sequencing run, such as insertions and deletions (INDELS) and base substitutions.
The ideal solution to minimize noise in downstream data analysis is to fit all samples that are to be compared into one sequencing run. Of course, this ideal is rarely possible, and many sequencing projects require multiple runs from which samples are compared. In this case as well, including spike-in controls and reference samples is vital. One step in the pre-sequencing workflow, namely library amplification by PCR, can be a key contributor of avoidable noise. For library amplification PCR, less really is more. The amount of technical noise increases the more a library is amplified. Bear in mind, this library is ultimately diluted significantly before being sequenced. In practice, one obtains better data from libraries that have undergone fewer PCR cycles. This is also why over-cycling can be a serious issue even with Unique Molecular Identifiers (UMIs) present. A future chapter of Lexicon will dive into PCR amplification in greater detail, and we will also introduce UMIs.
What we do here at Lexogen is design our protocols with the highest priority on output integrity. Put another way, Lexogen wants to make sure your data is as good as it can possibly be. One way we do this is by minimizing the disparity between the amount of library required for sequencing and the amount of post-PCR product, thus minimizing the technical noise introduced by the “random drawing” effect. In practice, this means that we are not focused on maximizing the ultimate yield of the library, since doing so would be counter-productive in the pursuit of excellent RNA sequencing performance. Instead, we focus on producing what is ultimately more than enough to sequence, while maximizing data quality.
Are you ready to become an RNA Expert?
Sign up and gain access to helpful checklists in PDF format that can assist you in your experiments. In addition, you’ll have the opportunity to download the RNA LEXICON E-BOOK in PDF format as well.





