SPLIT RNA Extraction Kit
- High-quality RNA for all downstream applications
- Efficient extraction of total RNA, including small RNA
- Universal – species-independent
- Fast and convenient protocol – extracted RNA in 30 minutes

Description
SPLIT RNA Extraction Kit
The SPLIT RNA Extraction Kit enables fast and highly efficient extraction of high-quality, high-purity RNA from various biological samples, including cell culture, animal and plant tissue, and fluid samples. The obtained RNA is ideal for seamless library preparation for Next Generation Sequencing and other demanding applications such as full-length reverse transcription, sample preparation for microarray analysis, or RT-qPCR. SPLIT recovers the complete RNA size range, including small RNAs (<200 nt). Additionally, large and small RNA-enriched fractions can be extracted by following a supplemental protocol.
High yields of high-quality RNA
RNA extracted with the SPLIT RNA Extraction Kit has a high RIN quality score for all types of samples. A RIN of 10 and a 28S / 18S rRNA ratio of 2.7 can be obtained from cell culture. Extractions from tissue samples usually result in RNA with a RIN of 8.0 – 9.5 (Figure 1). SPLIT RNA Extraction Kit extracts RNA with high efficiency. The typical yield of extracted RNA from mouse liver ranges from 4.0 – 4.5 µg total RNA / mg tissue.
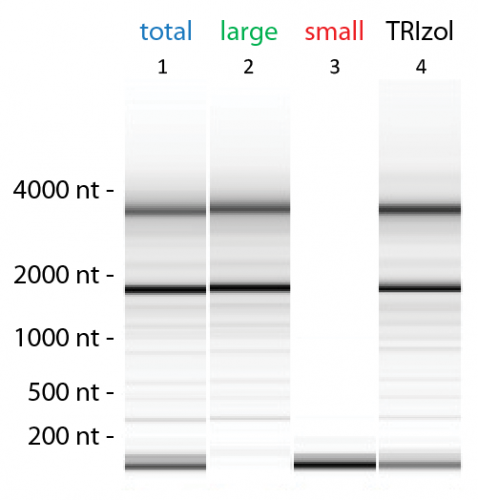
Small and Large RNA-enriched Fractions
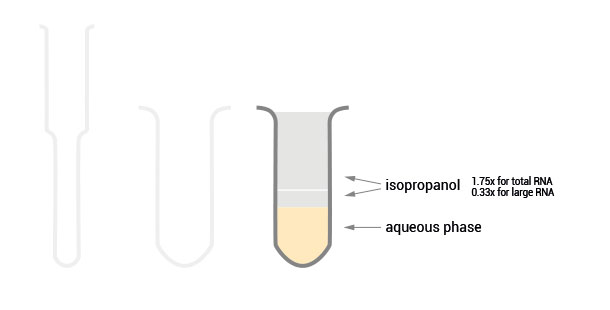
The SPLIT Kit can be used for the extraction of either total RNA (<17 nt to >10,000 nt) or for the isolation of the large RNA-enriched fraction (cutoff at ~150 nt), with the option to obtain the small RNA-enriched fraction separately (Figure 1).Universal, Species-independent RNA Extraction
SPLIT RNA Extraction Kit can be used with a broad range of material. A protocol is provided for animal and plant tissue, cell cultures, liquid samples, and FFPE samples.Total RNA, Including miRNAs
SPLIT RNA Extraction Kit effectively extracts RNA of all size range, including small RNAs (≥17 nt). Efficient recovery of siRNAs and miRNAs down to 17 nt in total RNA or the small RNA-enriched fraction has been shown in spike-in experiments with small RNA markers (Figure 2).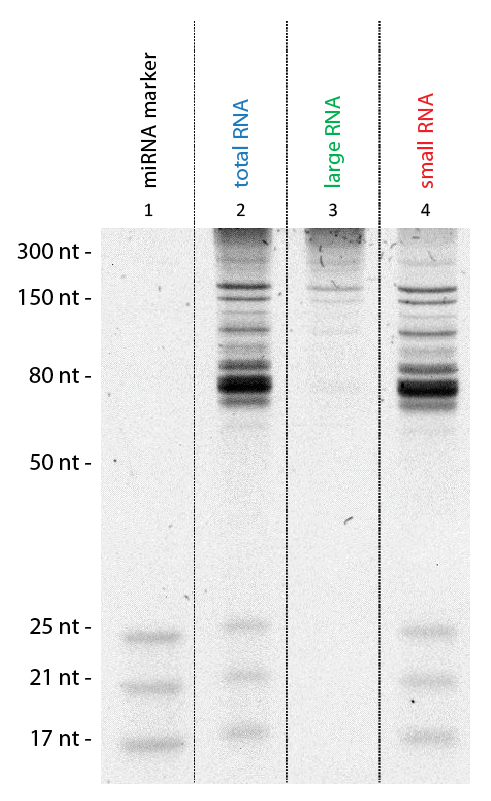
Fast and Convenient Protocol
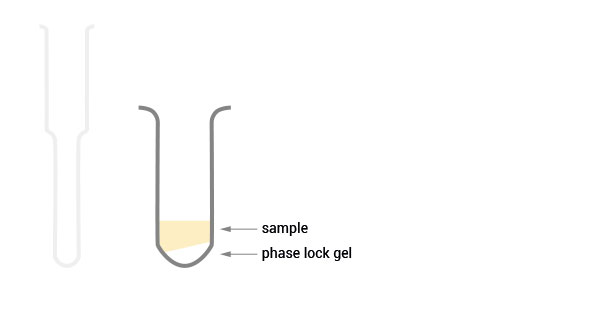
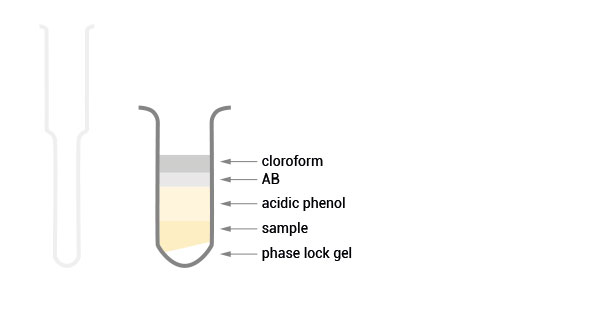
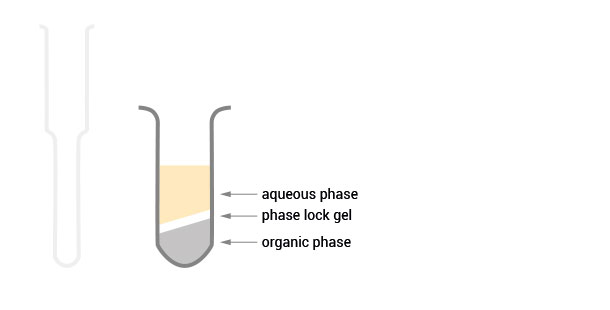
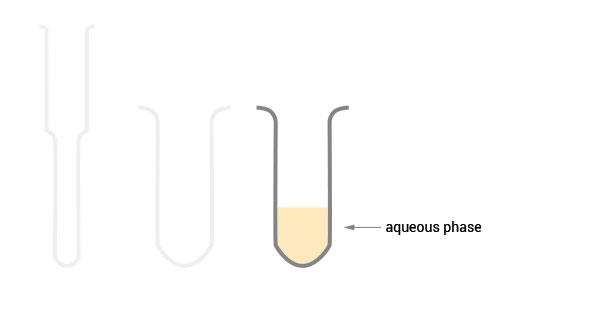
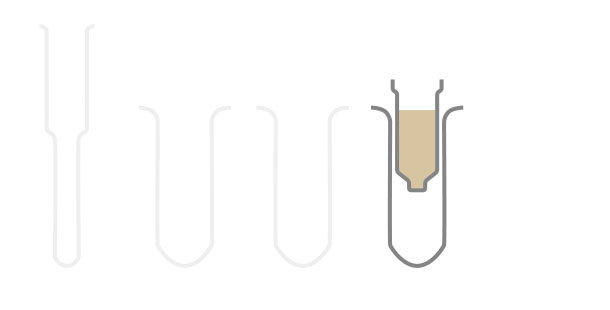
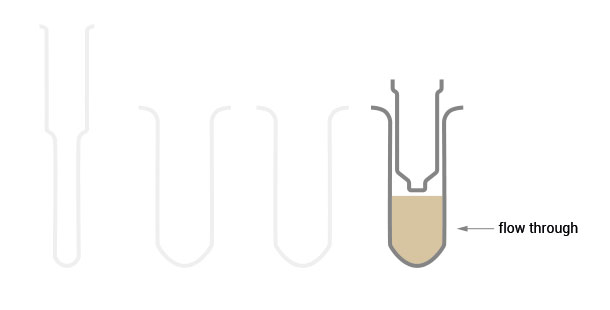
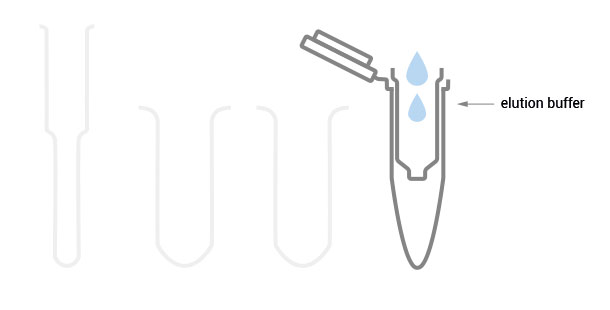
RNA can be extracted within 30 minutes. SPLIT RNA Extraction Kit contains Phase Lock Gel tubes allowing fast, comfortable, and safe phase separation.Workflow
The SPLIT RNA Extraction Kit contains reagents for the isolation of total RNA or the large RNA-enriched fraction from 48 samples, or small and large RNA-enriched fractions from 24 samples.
30 min
Featured Publications
FAQ
Frequently Asked Questions
Access our frequently asked question (FAQ) resources via the buttons below.
Please also check our General Guidelines and FAQ resources!
How do you like the new online FAQ resource? Please share your feedback with us!
Troubleshooting Guide
View the Troubleshooting Guide
| Problem | Likely cause | Comments and suggestions |
|---|---|---|
| No or poor phase separation | Cooled phase-lock gel or insufficient centrifugation |
|
| Incorrect cut-off | Volume of isopropanol addition |
|
| Degraded RNA | RNA source |
|
| RNase contamination |
| |
| Low absorption ratios (i.e., peak at 230 nm) | Contamination with organic solvents, salts, or metal ions |
|
| Low or no RNA yield | RNA remains on spin column |
|
| Spin column was overloaded |
| |
| Problem in downstream application | Salt or ethanol carry-over during elution |
|
Downloads
SPLIT RNA Extraction Kit
Safety Data Sheet
If you need more information about our products, please contact us through support@lexogen.com or directly under +43 1 345 1212-41.
First time user of SPLIT?
First Time User? We’re excited to offer you an exclusive introductory offer.
Buy from our webstore
Need a web quote?
You can generate a web quote by Register or Login to your account. In the account settings please fill in your billing and shipping address. Add products to your cart, view cart and click the “Generate Quote” button. A quote in PDF format will be generated and ready to download. You can use this PDF document to place an order by sending it directly to sales@lexogen.com.
Web quoting is not available for countries served by our distributors. Please contact your local distributor for a quote.