TeloPrime Full-Length cDNA Amplification Kit V2
- Full-length cDNA generation with high yield
- Exceptional 5‘ cap specificity
- Ideal for long-read NGS library generation
(e.g. PacBio™ and Oxford Nanopore™) - 1 ng – 2 μg total RNA input
- Flexible protocol enables use of custom primers
for reverse transcription
Description
TeloPrime Full-Length cDNA Amplification Kit V2
The TeloPrime Full-Length cDNA Amplification Kit V2 is an all-in-one protocol for generating full-length cDNA from total RNA.
Full-Length cDNA Synthesis
Based on Lexogen´s unique Cap-Dependent Linker Ligation (CDLL) and long reverse transcription (long RT) technology, it is highly selective for full-length RNA molecules that are both capped and polyadenylated.
For Multiple Downstream Applications
The full-length cDNA products can be used for various downstream applications such as NGS, RACE, cloning, microarray probes, and normalization. It enables the detection and correct quantification of splice variants and their true transcription start- and end-sites, in both short and long mRNA molecules. For further full-length or gene-specific PCRs, Lexogen offers a TeloPrime PCR Add-on Kit V2 (Cat.No.018.16) containing 16 rxn. This kit contains the PCR Forward and Reverse Primer separately, hence they can be alternatively substituted with the gene-specific primer of interest.
Full-Length cDNA Synthesis
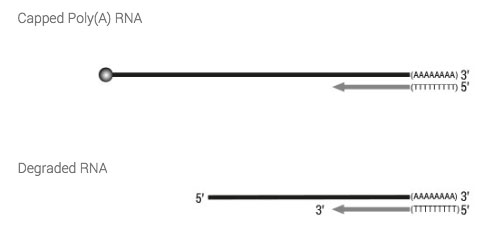
Based on the CDLL technology the ligation of the 5’ linker to the 3’ end of the cDNA takes place in a highly cap-dependent manner, providing an exceptional cap-specificity and eliminating cDNA products from degraded mRNA.
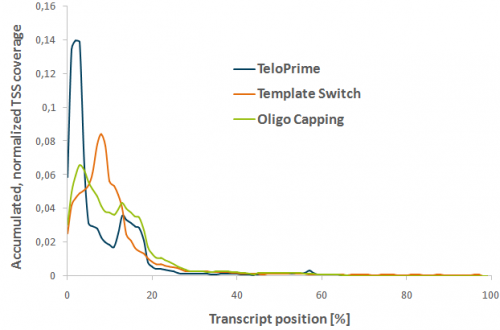
In an mRNA-Seq experiment, mRNAs were tagged at their 5’ end using TeloPrime, Template-Switch (TS) or Oligo Capping (OC). Transcript start sites (TSS) were mapped to the mouse genome. The accumulated TSS read coverage is plotted versus the normalized annotated transcript length to show relative TSS mapping for the top 500 expressed genes.
Workflow
The TeloPrime Full-Length cDNA Amplification Kit V2 is a protocol for generating full-length cDNA from total RNA. It is based on Lexogen´s unique Cap-Dependent Linker Ligation (CDLL) and long reverse transcription (long RT) technology, and is highly selective for full-length RNA molecules that are both capped and polyadenylated.
10-18 hrs
1 hr 25 min
Firstly, full-length cDNA synthesis is initiated by oligodT primed long reverse transcription. This helps to preserve the complete RNA sequence information in the cDNA before a cap selection is carried out. In addition a more stable RNA/cDNA hybrid is created that is maintained throughout post RT purification. This double-stranded (ds) hybrid is also important for the specificity of the subsequent Cap-Dependent Linker Ligation reaction.
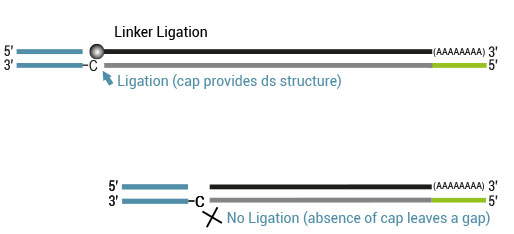
The double-stranded adapter with a 5’ C overhang allows for an atypical base-pairing with the inverted G of the cap structure. By using a ds-specific ligase, the ligation only takes place if the cap is present and if the RT has really reached the 5’ end of the mRNA. No ligation takes place if no cap is present e.g., in degraded RNA (low RIN) or if the RT has terminated prematurely because of secondary structures.
Based on this Cap-Dependent Linker Ligation only products with the 5’ tag get extended in the second-strand synthesis.
All remaining background is eliminated and the 5’ tagged full-length cDNA is converted into full-length ds cDNA.
Featured Publications
FAQ
Frequently Asked Questions
Access our frequently asked question (FAQ) resources via the buttons below.
Please also check our General Guidelines and FAQ resources!
Downloads
Safety Data Sheet
If you need more information about our products, please contact us through support@lexogen.com or directly under +43 1 345 1212-41.
First time user of TeloPrime?
First Time User? We’re excited to offer you an exclusive introductory offer.
Buy from our Webstore
Need a web quote?
You can generate a web quote by Register or Login to your account. In the account settings please fill in your billing and shipping address. Add products to your cart, view cart and click the “Generate Quote” button. A quote in PDF format will be generated and ready to download. You can use this PDF document to place an order by sending it directly to sales@lexogen.com.
Web quoting is not available for countries served by our distributors. Please contact your local distributor for a quote.