Poly(A) RNA Selection Kit V1.5
- Highly specific poly(A) enrichment
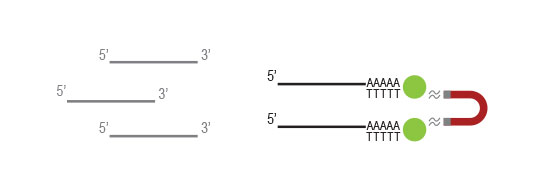
- Magnetic bead-based purification
- Various downstream applications such as RNA-Seq
- Fast workflow

Description
Poly(A) RNA Selection Kit V1.5
The Poly(A) RNA Selection Module enables the rapid and highly specific enrichment of polyadenylated RNAs from total RNA samples.
Optimized V1.5 Upgrade
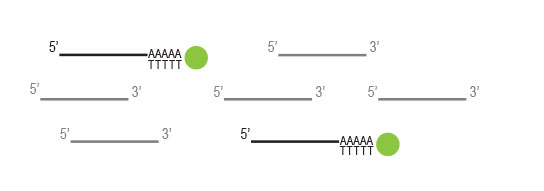
The upgrade V1.5 contains new oligo(dT) Magnetic Beads (MB), compared to the V1.0 kit. The beads and solutions included in the V1.5 kit offer improved performance.Highly Specific for Poly(A) RNA
Other RNA species (rRNA and tRNA) do not contain poly(A) sequences and therefore will not bind to the oligo(dT) beads.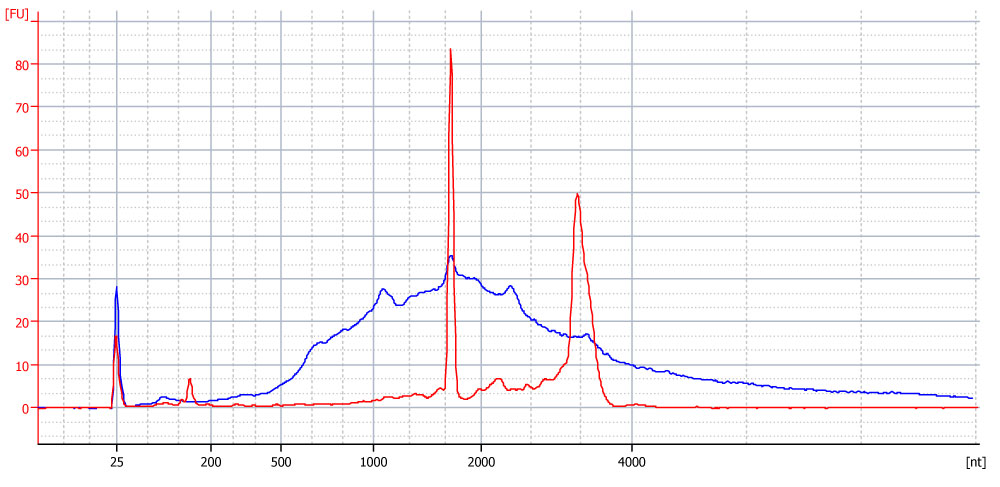
Various Downstream Applications
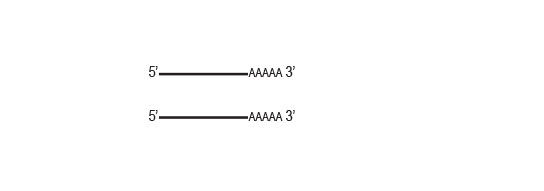
Isolated mRNA can directly be used for RNA-Seq library preparation (e.g., CORALL RNA-Seq Library Prep kits), SAGE, CAGE, cloning, microarrays, cDNA synthesis, and others.Rapid Turnaround
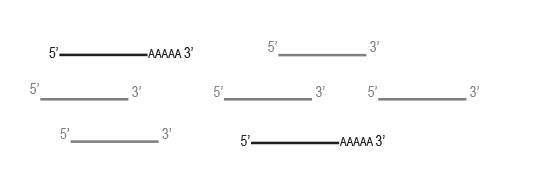
Polyadenylated RNAs can be isolated from total RNA samples within about one hour.Workflow
Any RNA without poly(A) stretches, such as rRNA
and tRNA,will not be captured by the oligodT beads
and will be washed away.
FAQ
Frequently Asked Questions
Access our frequently asked question (FAQ) resources via the buttons below.
Please also check our General Guidelines and FAQ resources!
How do you like the new online FAQ resource? Please share your feedback with us!
Downloads
Safety Data Sheet
If you need more information about our products, please contact us through support@lexogen.com or directly under +43 1 345 1212-41.
Buy from our webstore
Need a web quote?
You can generate a web quote by Register or Login to your account. In the account settings please fill in your billing and shipping address. Add products to your cart, view cart and click the “Generate Quote” button. A quote in PDF format will be generated and ready to download. You can use this PDF document to place an order by sending it directly to sales@lexogen.com.
Web quoting is not available for countries served by our distributors. Please contact your local distributor for a quote.