Reliable and most cost-efficient solution for gene expression analysis
– QuantSeq FWD
Do you have any questions?
Find the ideal kit for your application:
QuantSeq 3’ mRNA-Seq V2 Library Prep Kit with UDI
Straightforward expression profiling with QuantSeq 3’ mRNA-Seq gene expression kits: the cost-saving 3’ sequencing approach enables mRNA-focused readout and exceptional scalability.
QuantSeq FWD V2 with UDI generates sequencing-ready libraries from total RNA in <4.5 hours without the need for any pre-processing steps and presents the ideal choice for cost-efficient gene expression profiling studies of eukaryotic samples.
QuantSeq kits contain all reagents to complete RNA-Seq library preparation, including library generation reagents, library amplification reagents, Unique Dual Indices (UDI), and purification beads and solutions.
Add-on kits:
- PCR Add-on Kit for Illumina (Cat. No. 208)
- UMI Second Strand Synthesis Module for QuantSeq FWD (Cat. No. 081)
- Globin Block Modules for QuantSeq (homo sapiens Cat. No. 070, sus scrofa Cat. No. 071) for blocking of globin mRNA
- BC1 Block Module for QuantSeq (Cat. No. 167) for blocking of abundant BC1 transcripts in rodent brains
System compatibility:
- Illumina (directly compatible)
- Element AVITI (directly compatable with Cloudbreak Freestyle, or ADEPT)
- MGI (compatible after conversion)
Performance
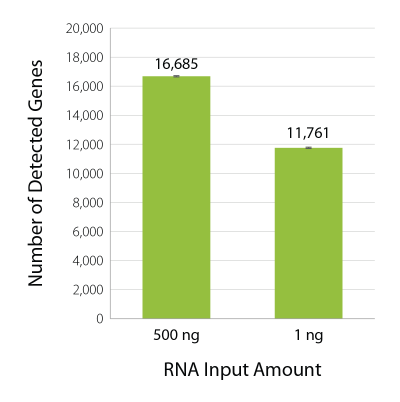
Excellent Gene Detection Across a Wide Range of Input Amounts
The majority of reads generated by QuantSeq library preparation from 500 ng or 1 ng RNA present uniquely mapping reads (Table 1). QuantSeq 3′ mRNA-Seq V2 also provides excellent gene detection across a wide range of input RNA amounts and is thus ideal for gene expression studies (Fig. 1).
| QuantSeq 3' mRNA-Seq FWD V2 | ||||
|---|---|---|---|---|
| Input RNA | 500 ng | 1 ng | ||
| Mapped Reads | 99.45% | 99.12% | ||
| Uniquely Mapped Reads | 89.77% | 88.70% | ||
| Multimapping Reads | 9.67% | 10.42% | ||
| Reads Mapped to rRNA* | 0.29% | 1.45% | ||
| Reads Mapped to Protein-coding Genes | 93.71% | 88.72% | ||
| *Reads mapping to cytoplasmic rRNA excluding polyadenylated mitochondrial rRNA transcripts. | ||||
High Reproducibility Across Various Inputs
Transcript Coverage of 3' Ends for Cost-efficient Sequencing
Performance on FFPE Samples
QuantSeq FWD V2 with UDI shows consistent correlation of gene detection between mouse spleen samples derived from fresh frozen (FF) and FFPE tissue at various gene detection thresholds (Fig. 4)
For optimized performance on FFPE samples, QuantSeq FFPE kits are recommended. QuantSeq FFPE is validated and QC’ed on FFPE material and seamlessly integrates with SPLIT One-step FFPE RNA Extraction kits for a fully validated workflow.
Figure 4 | Overlap of detected genes between fresh frozen (FF) and FFPE samples from 10 ng input (different mouse spleen samples) at 1M reads / sample. Data is shown for 1 CPM, 5 CPM, and 10 CPM (from left to right). RIN values: 3.7 (FF) and 2.9 (FFPE), DV200 values: 90% (FF) and 9% (FFPE).
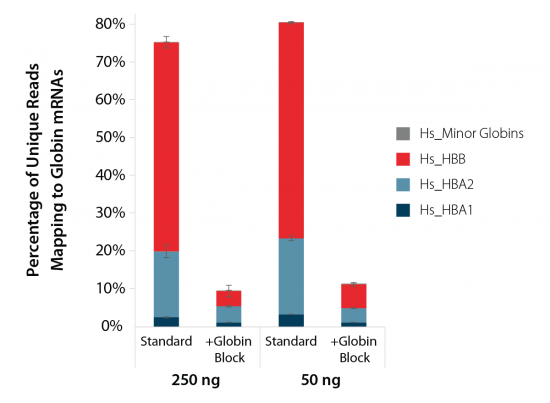
Performance on Blood Samples
QuantSeq is ideal for gene expression studies from whole blood. Libraries prepared with QuantSeq and Globin Block Modules show significant reduction of reads mapped to globin with depletion rates >80 % (Fig. 5). Lexogen RNA-Seq workflows for blood samples are compatible with established blood collection systems, including PAXgene™ Blood RNA Tubes.
Find out more in Publications using QuantSeq with Globin Block.
Deplete Abundant Transcripts in Rodent Brain Samples with BC1 Block and QuantSeq
The BC1 Block Module (RS-BC1 Block) for QuantSeq prevents the generation of library fragments from the abundant BC1 transcripts that are present in rodent brain samples by blocking their extension during second strand synthesis (Fig. 6). As a result, Reads mapping to BC1 are reduced and the number of uniquely mapping reads is increased (Table 2).
| without BC1 Block | with BC1 Block | |
|---|---|---|
| Mapped Reads | 97.60% | 98.00% |
| Uniquely Mapped Reads | 62.00% | 73.30% |
| Reads Mapped to BC1 | 12.70% | 0.01% |
| Reads Mapped to BC1 Homologs | 6.50% | 0.01% |
Performance on Plants
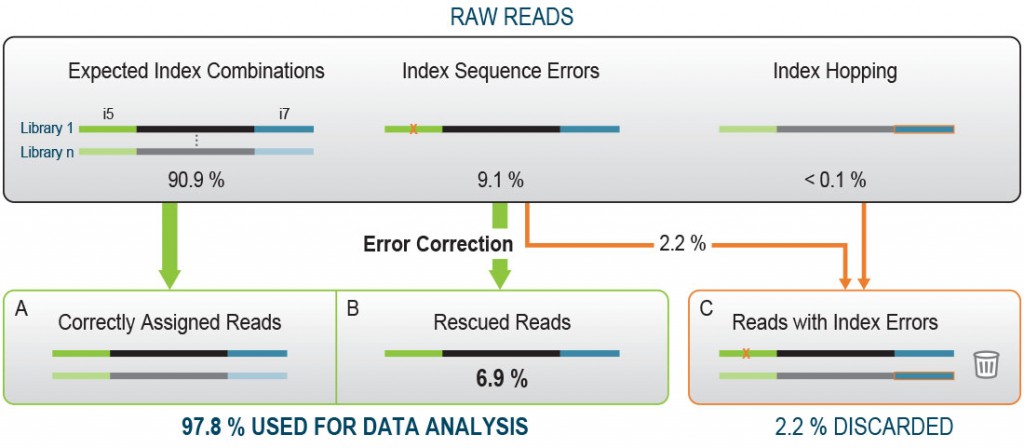
Optimal UDI deduplication capabilities
Lexogen’s patented 12 nt UDI allow for particularly high error correction rates, with optimally designed sequence-Levenshtein values. With Lexogen, you do not have to worry about lost reads or even misassigned reads! And you can benefit from our design even on libraries generated with other vendors’ kits since our UDI kits can adapt to most of the commercially available library generation solutions. For optimal demultiplexing and read rescue, we recommend Lexogen’s iDemux command line.
Please consult with us for more information: support@lexogen.com.
Figure 3 | Lexogen’s 12 nt design allows rescuing the majority of undetermined reads thanks to superior error correction features, thereby saving precious data (based on 96 pooled libraries; Illumina NextSeq500 run, demultiplexed with iDemux).
Workflow
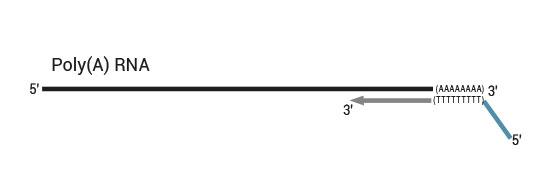
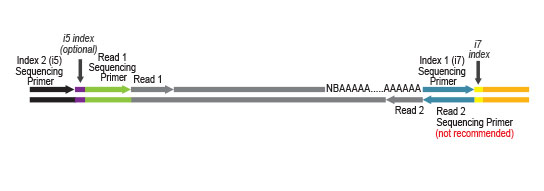
Library generation starts with oligodT priming containing the Illumina-specific Read 2 linker sequence.
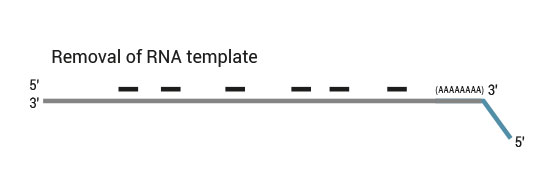
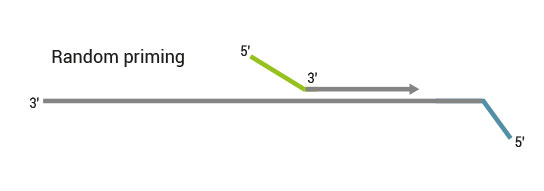
Second strand synthesis is initiated by random priming and a DNA polymerase. The random primer contains the Illumina-specific Read 1 linker sequence. At this step Unique Molecular Identifiers (UMIs) can be introduced by exchanging the Second Strand Synthesis Mix 1 (SS1) from the standard QuantSeq FWD Kit with UMI Second Strand Synthesis Mix (USS).
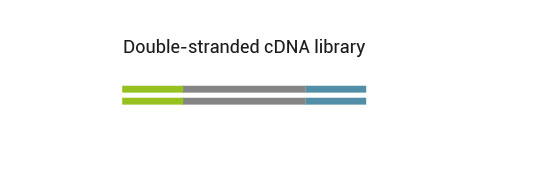
No purification is required between first and second strand synthesis. Second strand synthesis is followed by a magnetic bead-based purification step rendering the protocol compatible with automation.
NGS reads are generated towards the poly(A) tail and directly correspond to the mRNA sequence. To pinpoint the exact 3’ end, longer reads may be required (SR50, SR100, SR150). Although paired-end sequencing is possible, we do not recommend it for QuantSeq FWD. Read 2 would start with the poly(T) stretch, and as a result of sequencing through the homopolymer stretch, the quality of Read 2 would be very low.
Automation
Automated QuantSeq protocol for 3’ mRNA-Seq Library Preparation
Automating the process of library preparation has the advantage of avoiding sample tracking errors, dramatically increasing throughput, and saving hands-on time.
QuantSeq library preparation and has been successfully implemented on several liquid handlers:
- Perkin Elmer: Sciclone® / Zephyr®
- Hamilton: Microlab STAR / STARlet
- Agilent: NGS Workstation (NGS Bravo Option B)
- Beckman Coulter: Biomek FXP, Biomek i5, Biomek i7
- Eppendorf: EpMotion® 5075
- Opentrons® OT-2
- Tecan: DreamPrep® NGS
QuantSeq automation on other platforms may also be possible. Please contact support team for more information.
Considerations for Automation
Two key parameters must always be considered when automating a protocol:
- volume optimization
- script compatibility
In some instances, our kits will provide enough reagents while, in other instances, you will need a higher reagent volume or script adjustments. At Lexogen, we will help you find the best solution, tailored to your needs.
Before starting any new project involving automation, please reach out to our experts at support@lexogen.com
Please, also consider liaising with the robotic platform support team – they will be able to share the most recent version of the script.
Lexogen gladly supports the implementation of Lexogen-manufactured kits on liquid handlers, but not hardware or software issues linked to the original liquid handling instrument supplier.
Related resources:
FAQ
Frequently Asked Questions
Access our frequently asked question (FAQ) resources via the buttons below.
Please also check our General Guidelines and FAQ resources!
How do you like the new online FAQ resource? Please share your feedback with us!
Downloads
Safety Data Sheet
If you need more information about our products, please contact us through support@lexogen.com or directly under +43 1 345 1212-41.
Ordering Information
| Cat. № | Product Name | |||
|---|---|---|---|---|
| Full library preparation kits | Full library preparation kits | Full library preparation kits | ||
| 191.24 | QuantSeq 3’ mRNA-Seq V2 Library Prep Kit FWD with UDI 12 nt Set A1, 24 preps | |||
| 192.24 | QuantSeq 3’ mRNA-Seq V2 Library Prep Kit FWD with UDI 12 nt Set B1, 24 preps | |||
| 191.96 | QuantSeq 3’ mRNA-Seq V2 Library Prep Kit FWD with UDI 12 nt Set A1, 96 preps | |||
| 192.96 | QuantSeq 3’ mRNA-Seq V2 Library Prep Kit FWD with UDI 12 nt Set B1, 96 preps | |||
| 193.384 | QuantSeq 3’ mRNA-Seq V2 Library Prep Kit FWD with UDI 12 nt Sets A1 to A4, 384 preps | |||
| 194.96 | QuantSeq 3’ mRNA-Seq V2 Library Prep Kit FWD with UDI 12 nt Set A2, 96 preps | |||
| 195.96 | QuantSeq 3’ mRNA-Seq V2 Library Prep Kit FWD with UDI 12 nt Set A3, 96 preps | |||
| 196.96 | QuantSeq 3’ mRNA-Seq V2 Library Prep Kit FWD with UDI 12 nt Set A4, 96 preps | |||
| UDI Add-on Kits (UDI plate and library amplification module) | ||||
| 198.96 | Lexogen UDI 12 nt Unique Dual Indexing V2 Add-on Kit Set A1, 96 preps | |||
| 199.96 | Lexogen UDI 12 nt Unique Dual Indexing V2 Add-on Kit Set A2, 96 preps | |||
| 200.96 | Lexogen UDI 12 nt Unique Dual Indexing V2 Add-on Kit Set A3, 96 preps | |||
| 201.96 | Lexogen UDI 12 nt Unique Dual Indexing V2 Add-on Kit Set A4, 96 preps | |||
| 202.96 | Lexogen UDI 12 nt Unique Dual Indexing V2 Add-on Kit Set B1, 96 preps | |||
| 203.384 | Lexogen UDI 12 nt Unique Dual Indexing V2 Add-on Kit Sets A1 to A4, 384 preps | |||
First time user of QuantSeq?
First Time User? We’re excited to offer you an exclusive introductory offer.
Buy from our Webstore
Need a web quote?
You can generate a web quote by Register or Login to your account. In the account settings please fill in your billing and shipping address. Add products to your cart, view cart and click the “Generate Quote” button. A quote in PDF format will be generated and ready to download. You can use this PDF document to place an order by sending it directly to sales@lexogen.com.
Web quoting is not available for countries served by our distributors. Please contact your local distributor for a quote.