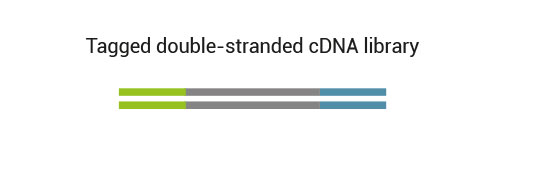
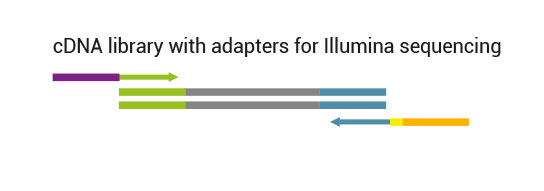
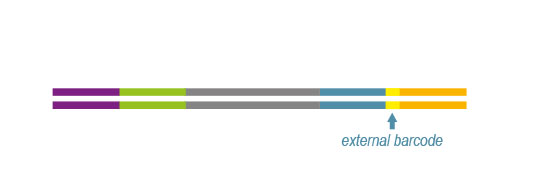
Custom-designed and flexible solution for targeted RNA-Seq
– QuantSeq-Flex
Do you have any questions?
Find the ideal kit for your application:
QuantSeq-Flex Targeted RNA-Seq Library Prep Kit V2
The QuantSeq-Flex Kit is a library preparation protocol designed to make Illumina compatible libraries from any RNA sample using custom primers. It is open for development by advanced RNA-Seq users based on their custom needs.
The kit is based on the QuantSeq FWD Kit for Illumina strategy, with compatible reagents and Read 1 linker sequence introduced by the second strand synthesis primer. Hence the reads generated during the NGS run directly correspond to the RNA sequence.
QuantSeq Flex can be ideally combined with our QuantSeq with UDI V2 kits, allowing for optimal library multiplexing with Lexogen’s patented 12 nt Unique Dual Indices (Cat. No. 191 to 196).
QuantSeq-Flex Kit provides modules for Reverse Transcription (RT) and SSS, allowing users to choose their own primers for either one of these steps or both of them. This offers maximum flexibility in the choice of targets and input material.
With this highly flexible QuantSeq kit the following types of libraries can be generated:
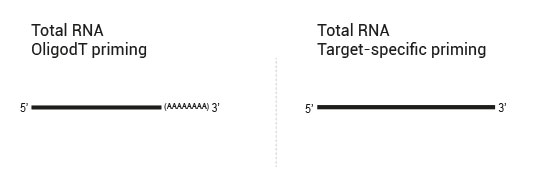
- OligodT primed in reverse transcription, random primed in second strand synthesis (QuantSeq 3’ mRNA-Seq)
- OligodT primed in reverse transcription, target-specifically primed in second strand synthesis (targeted 3’ mRNA-Seq)
- Target-specifically primed in reverse transcription, random primed in second strand synthesis (targeted RNA-Seq, allows for identification of novel fusions)
- Target-specifically primed in reverse transcription, target-specifically primed in second strand synthesis (targeted RNA-Seq, known targets detectable only)
Whether it is gene expression analysis, targeted sequencing, adapter ligation-based RNA-Seq or any other experiment where a certain RNA region needs to be sequenced, QuantSeq-Flex can be used together with user-supplied primers.
Performance
Direct Counting for Gene Expression Quantification
Just one fragment per transcript is produced; therefore no length normalization is required. This allows more accurate determination of gene expression values and makes QuantSeq the best alternative to microarrays and conventional RNA-Seq in gene expression and eQTL studies.
Rapid Turnaround
Simple workflow allows generating libraries ready for sequencing within 4.5 hours. This already includes hands-on time, which is less than 2 hours.
Cost Saving Multiplexing
QuantSeq libraries are intended for a high degree of multiplexing. At Lexogen, we offer you different options:
- Combinatorial Dual Indexing (CDI): up to 9,216 samples can be multiplexed/lane on an Illumina flow cell using the 96 i7 6nt indices (included in the original QuantSeq kit) together with the 96 i5 indices as part of the Lexogen i5 6 nt Dual Indexing Add-on Kit (Cat. No. 047).
- Unique Dual Indexing (UDI): up to 384 samples can be multiplexed/lane on an Illumina flow cell using Lexogen’s patented 12 nt UDI sets – with exceptional error correction capabilities, therefore rescuing most of your reads. For this option, please consider our QuantSeq with UDI V2 kits (Cat. No. 191 to 196).
In order to be compatible with the supplied indices (barcode primers) please note our recommendations for the custom primer design (see Appendix D: Primer Design, QuantSeq-Flex User Guide, p.28)
For more information about indexing strategies offered by Lexogen.
An RNA-Seq Experiment Tailored to Your Needs
QuantSeq-Flex modular kit gives maximum flexibility to the user encouraging implementation of RNA-Seq as a part of the experimental pipeline. One fragment per transcript produced and usage of custom primers makes this kit a perfect solution for cost-saving high-throughput gene expression analysis. QuantSeq-Flex is suitable for targeted sequencing of gene panels, enrichment, detection of fusion genes, and various other types of transcripts.
Workflow
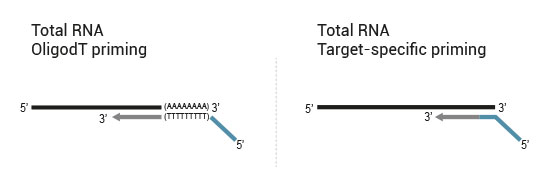
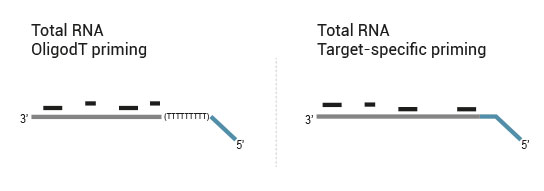
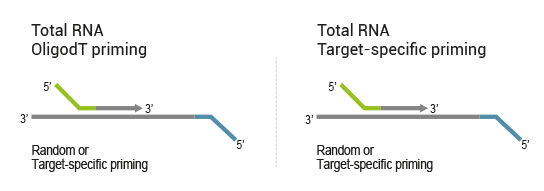
Library generation starts either with oligodT (included in the kit) or a target-specific primer containing sequencing platform - compatible linker sequences.
Second strand synthesis is initiated by random (included in the kit) or target-specific priming and a DNA polymerase. The primers also contain sequencing platform - compatible linker sequences.
No purification is required between first and second strand synthesis. Second strand synthesis is followed by a magnetic bead-based purification step rendering the protocol compatible with automation.
Featured Publications
Automation
Automated QuantSeq protocol for 3’ mRNA-Seq Library Preparation
Automating the process of library preparation has the advantage of avoiding sample tracking errors, dramatically increasing throughput, and saving hands-on time.
QuantSeq library preparation and has been successfully implemented on several liquid handlers:
- Perkin Elmer: Sciclone® / Zephyr®
- Hamilton: Microlab STAR / STARlet
- Agilent: NGS Workstation (NGS Bravo Option B)
- Beckman Coulter: Biomek FXP, Biomek i5, Biomek i7
- Eppendorf: EpMotion® 5075
- Opentrons® OT-2
QuantSeq automation on other platforms may also be possible. Please contact support team for more information.
Two key parameters must always be considered when automating a protocol:
- volume optimization
- script compatibility
In some instances, our kits will provide enough reagents while, in other instances, you will need a higher reagent volume or script adjustments. At Lexogen, we will help you find the best solution, tailored to your needs.
Before starting any new project involving automation, please reach out to our experts at support@lexogen.com
Please, also consider liaising with the robotic platform support team – they will be able to share the most recent version of the script.
Lexogen gladly supports the implementation of Lexogen-manufactured kits on liquid handlers, but not hardware or software issues linked to the original liquid handling instrument supplier.
Related resources:
FAQ
Frequently Asked Questions
Access our frequently asked question (FAQ) resources via the buttons below.
Please also check our General Guidelines and FAQ resources!
How do you like the new online FAQ resource? Please share your feedback with us!
Downloads
QuantSeq-Flex Targeted RNA-Seq Library Prep Kit V2 for Illumina platforms
autoQuantSeq 3′ mRNA-Seq Library Prep Kit for Illumina
Agilent Bravo – Please inquire at info@lexogen.com for the automation scripts
Safety Data Sheet
If you need more information about our products, please contact us through support@lexogen.com or directly under +43 1 345 1212-41.
Want to try QuantSeq-Flex Targeted RNA-Seq V2 on your samples?
Dear Customer,
If you are interested in discussing your QuantSeq Flex project with Lexogen scientist, please contact us at info@lexogen.com.
Buy from our Webstore
Need a web quote?
You can generate a web quote by Register or Login to your account. In the account settings please fill in your billing and shipping address. Add products to your cart, view cart and click the “Generate Quote” button. A quote in PDF format will be generated and ready to download. You can use this PDF document to place an order by sending it directly to sales@lexogen.com.
Web quoting is not available for countries served by our distributors. Please contact your local distributor for a quote.