Experience robust and reliable results with QuantSeq
Do you have any questions?
Find the ideal kit for your application:
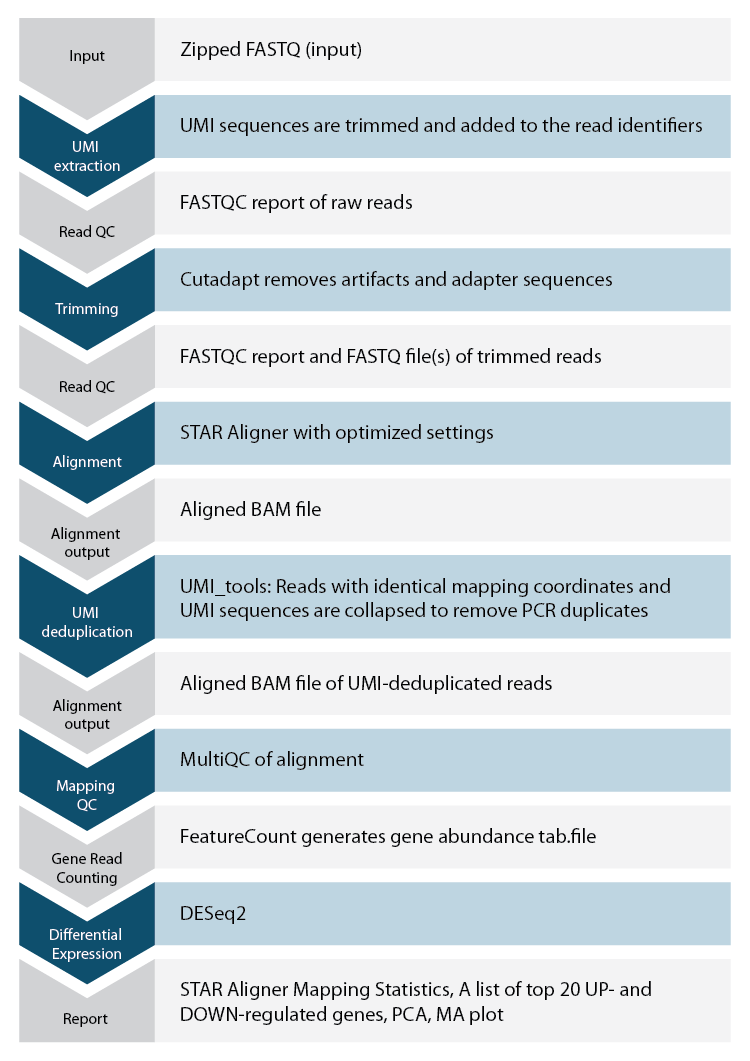
Lexogen’s Automated Data Analysis Solution
Lexogen offers an automated solution for QuantSeq data analysis based on state-of-the-art Lexogen’s proprietary pipeline. Thus, every user, even without bioinformatics experience, can analyze QuantSeq samples in a convenient and fast way.
Each QuantSeq FWD and REV kit is provided with codes for free data analysis on Lexogen’s data analysis solution, including differential expression analysis. Data analysis codes for additional runs or bigger files can be purchased from Lexogen. Please contact us at sales@lexogen.com.
The QuantSeq data analysis pipeline has been implemented in the Partek Flow software and is available for any Partek Flow user at www.partek.com/lexogen-quantseq-pipeline.
The QuantSeq pipeline is available on the OnRamp Platform ROSALIND, which allows Lexogen’s customers to analyze, interpret and collaborate globally on differential gene expression analysis without the need for specialized bioinformatics or programming skills.
QuantSeq data analysis pipeline is also available on the open-source data analysis software Chipster. For more information, you can watch a tutorial video.