SIRVs
The most comprehensive
RNA spike-in controls
SIRV Sets
SIRV Set Selection Guide
The Spike-In RNA Variants of the isoform, the ERCC, and the long SIRV modules (see Modular Design) are realized as defined mixes, and for some modules, different mixes are available. Mixes, and combinations thereof, are available in the form of SIRV sets.
We use the following definitions:
| Module | Group of spike-in controls that mimic predominantly one aspect of transcriptome complexity. |
|---|---|
| Mix | SIRVs of the same module that are combined in precise defined molarity. |
| Set | Term for the combination of mixes or modules. |
Table 1 presents a SIRV set selection guide to help you finding the right set of modules and mixes for your application.
| SIRV-Set 1 | SIRV-Set 2 | SIRV-Set 3 | SIRV-Set 4 | |||
|---|---|---|---|---|---|---|
| Cat. No | 025.03 | 050.0* | 051.0* | 141.0* | ||
| Module(s) | Isoforms | Isoform Mixes E0, E1, E2 | Isoform Mix E0 | Isoform Mix E0 | Isoform Mix E0 | |
| ERCC | X | X | ERCC Mix 1 | ERCC Mix 1 | ||
| long SIRVs | X | X | X | long SIRVs | ||
| Property | Isoform detection, and quantification | ✓ | ✓ | ✓ | ✓ | |
| Dynamic range | partially | X | ✓ | ✓ | ||
| Length > 2.5 kb | X | X | X | ✓ | ||
| Pipeline Validation | ✓ | partially | partially | partially | ||
| Sample Control | X | ✓ | ✓ | ✓ | ||
| Number of spike-in transcripts in each mix | 69 (69 isoforms in each Mix) | 69 (69 isoforms) | 161 (69 isoforms, 92 ERCCs) | 176 (69 isoforms, 92 ERCCs, 15 long SIRV) | ||
SIRV-Set 1 (Cat. No 025) contains the isoform mixes E0, E1 and E2 of the isoform module, SIRV-Set 2 (Cat. No 050) provides the isoform Mix E0 only, SIRV-Set 3 (Cat. No 051) has the SIRV Isoform Mix E0 in a mixture with the ERCCs, and SIRV-Set 4 (Cat. No 141) is a mixture of the long SIRVs with SIRV Isoform Mix E0 and the ERCCs.
*Refers to number of vials, 1 or 3. The ERCC Module includes ERCC Mix 1 (Munro et al., 2014)
✓: applicable, X : not applicable, and partly applicable (or parts of the sets applicable).
For more information please consult the respective User Guides in the Downloads section.
SIRV-Set 1: Isoform Mixes E0, E1 and E2
SIRV-Set 2: Isoform Mix E0
The isoform Mix E0 is available on its own as SIRV-Set 2 (Cat. No 050) with all 69 isoforms being present at equimolar concentrations. Its applications include RNA-Seq experiments that need to be validated for the detection of a complex mixture of isoforms without applying a high read depth to cover transcripts at different concentrations. Among these are sequencing runs on long-read NGS platforms as provided by Oxford Nanopore Technologies and Pacific Biosciences. These can produce full-length reads to identify the isoforms faithfully. However, unlike short-read platforms they do not provide the millions of reads necessary to detect and quantify isoforms across a larger dynamic range, in particular if these spike-ins only constitute a very small fraction of the total RNA.
Deviations from the expected equimolar outcome can be quantified, which allows for evaluation of the performance of isoform-centered workflows. On the gene level, quantifications of the individual SIRV isoform can be summed up for each SIRV gene, which permits the validation of pipelines working with data stemming from individual isoforms but focused on gene expression calculations only.
SIRV-Set 2 is very suitable for the calculation of concordance, since the experiments’ fingerprints depends solely on their dealing with the SIRV isoform’s complexity but not on input concentration differences between these isoform transcripts.
Reads from SIRV-Set 2 can be downsampled to emulate data representing the situations in lower concentration ranges. A repeated mapping and assignment provides adequate measures for the RNA-Seq experiment ability to detect variants and measure its concentrations in a different band with. Using such iterative approach SIRV-Set 2 is capable to map the entire abundance spectrum.
SIRV-Set 3: Isoform Mix E0 & ERCCs
SIRV-Set 3 (Cat. No. 051) contains the isoform Mix E0 and the ERCC Mix 1 in equal shares. Both contribute equally to the final mass.
The mixture of 69 SIRV isoform transcripts and 92 non-overlapping ERCC RNAs addresses the need for complex spike-in RNA controls that cover both, a high level of isoform complexity and a large concentration range. Together, they enable an even more comprehensive quality assessment and monitoring across the whole RNA-Seq workflow to derive technical details and telling fingerprints for comparing individual samples, and experiments.
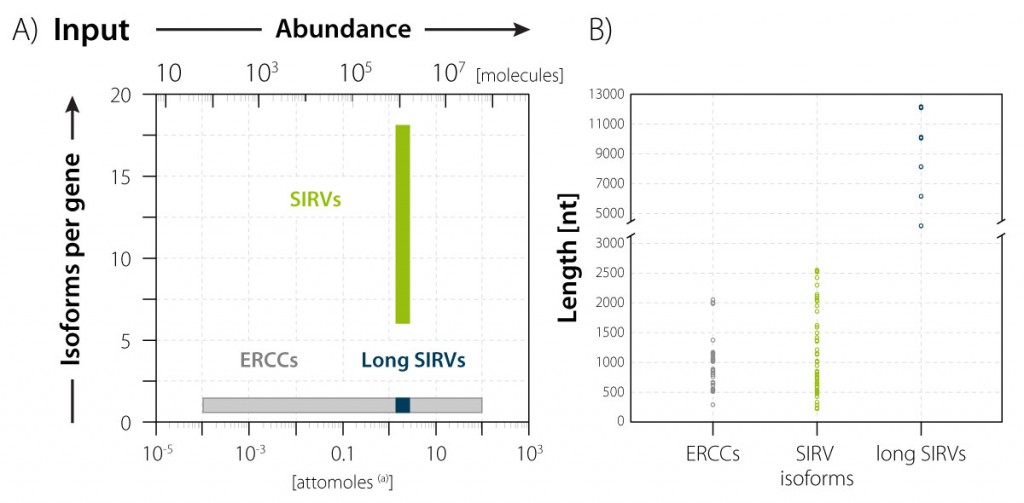
The single-isoform ERCC transcripts cover concentrations of 6 orders of magnitude and are complemented by the equimolar SIRV isoforms. Figure 2 illustrates this added dimension by showing the covered complexity plotted versus the input concentration.
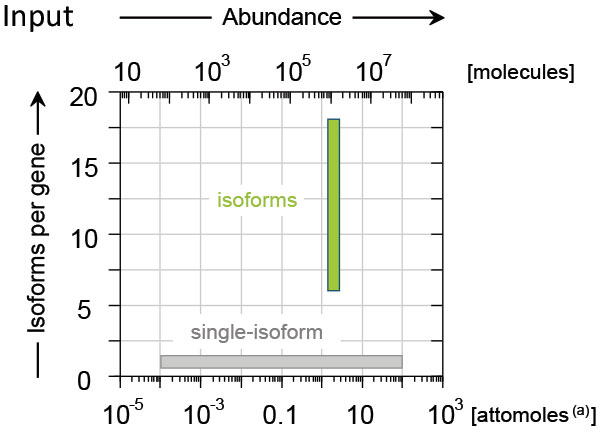
Figure 2 ǀ Concentrations and complexity of SIRV isoforms and single-isoform ERCC transcripts in SIRV-Set 3. Top; the isoform module with 69 transcripts in 7 gene loci contains all species at the same molarity (green bar). It covers the medium to high range of natural occurring isoform complexity. The single-isoform module with 92 ERCC transcripts covers concentrations of 6 orders of magnitude (grey bar), which is sufficient to represent the entire dynamic range of natural occurring transcripts. (a) The amount of attomoles refers to the typical amount that is spiked into 100 ng total RNA with the aim of attracting approx. 1% of the mRNA-Seq reads – subject to mRNA content and pipeline parameter for which, of course, the modules control for.
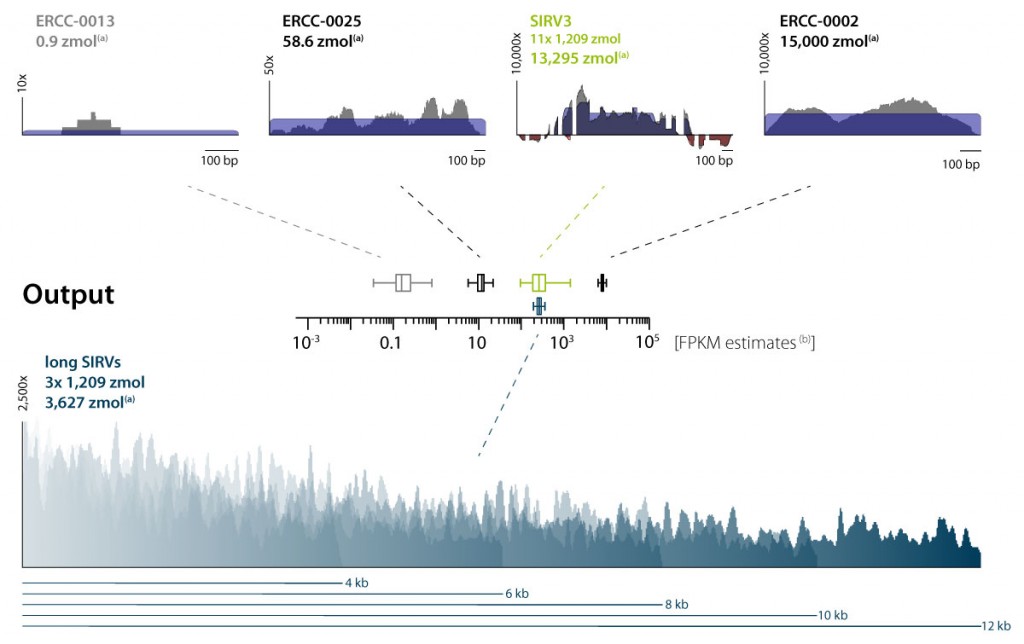
The accuracy (systematic error) and precision (random error) in quantifying single-isoform transcripts in RNA-Seq experiments is predominantly concentration- and read-depth dependent. While reads usually map uniquely to ERCC transcripts, the precision remains coverage dependent with reads following typically a Poisson distribution.
Isoform detection and quantification requires a sufficient coverage, and therefore the isoform spike-ins are added all at the same concentration and in the upper range of the ERCCs. Thereby, the issue of identifying a given isoform is not mingled up with differing concentrations. The overall higher input concentration allows for sufficient reads to be obtained in RNA-Seq experiments for isoform identification. Still, quantification of SIRV isoforms remains challenging on both short-read systems (due to assignment issues) and long-read platforms (e.g. because of per base error, low read numbers, and amplification bias). This is indicated in Figure 3 by a larger error margin for the isoforms than e.g. for ERCCs at an even lower concentration.
SIRV-Set 4: Isoform Mix E0, ERCCs, and long SIRVs
SIRV-Set 4 (Cat. No. 141) contains the long SIRV module with 15 RNAs of 4 – 12 kb length in addition to the 69 isoforms and 92 ERCC transcripts of SIRV-Set 3. The long SIRVs are present at concentrations identical to the SIRV isoforms, and therefore quantification is not affected by isoform complexity, resulting in a smaller standard error (Figure 3).
SIRV-Set 4 covers three spike-in aspects: isoform complexity, abundance, and length. These are clearly segregated with the long SIRVs having non-overlapping sequences and equal concentrations (Figure 4).