RiboCop rRNA Depletion Kit HMR plus Globin
- Depletion of all cytoplasmic and mitochondrial rRNA
- NEW! Combined rRNA and globin mRNA depletion
- NEW! Innovative probe design minimizing off-target effects
- 1 ng to 1 µg total RNA input (FFPE compatible)
RiboCop for Human/Mouse/Rat plus Globin
Total RNA from mammalian blood is comprised of highly abundant undesired RNA species, such as ribosomal RNA (rRNA), accounting for ~80 – 90 % of total RNA, and globin mRNA, representing 30 – 80 % of all mRNAs. Lexogen’s RiboCop rRNA Depletion Kit for Human/Mouse/Rat plus Globin removes undesired cytoplasmic (28S, 18S, 5.8S, and 45S rRNA) and mitochondrial rRNA (mt16S, mt12S, and 5S), and globin mRNA from intact as well as degraded whole blood RNA.
RiboCop for Human/Mouse/Rat plus Globin (HMR+Globin) is offered as a stand-alone kit (Cat. No. 145) or as bundled version with CORALL RNA-Seq V2 (Cat. No. 185 – 186). RiboCop Kits are compatible with all random primed total RNA-Seq library prep kits.
Performance
Sequence what matters most! RiboCop enables specific, highly effective, non-enzymatic depletion of ribosomal RNA from a broad range of sample types, including low input and degraded blood samples.
Globin Depletion Increases Gene Detection
RiboCop for Human/Mouse/Rat plus Globin (HMR+Globin) simultaneously depletes rRNA and globin mRNA, providing a highly convenient workflow for blood samples and freeing up sequencing space for RNAs of interest (Fig. 1). Thus, removal of globin mRNA leads to a significant increase in gene detection (Fig. 2).
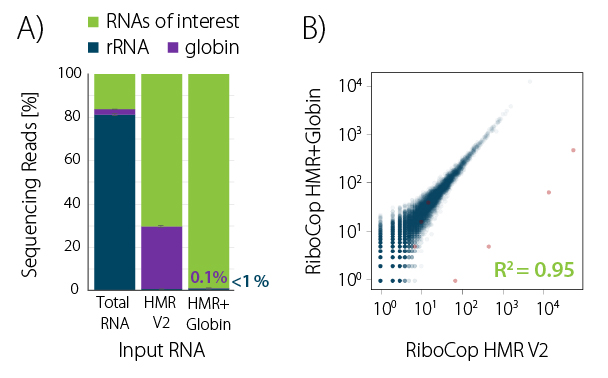
Figure 1 | RiboCop rRNA Depletion for Human/Mouse/ Rat plus Globin (HMR+Globin) efficiently removes rRNA and globin mRNA from human blood RNA. A) RNA was extracted from fresh human donor blood using SPLIT RNA Extraction. 10 ng whole blood RNA were depleted with RiboCop HMR V2 or HMR+Globin. Libraries were prepared with the CORALL Total RNA-Seq Library Prep Kit and sequenced on NextSeq500 (1×75 bp). Reads were mapped against the human reference genome (GRCh38.95) and reads mapping to rRNA (blue) and globin (purple) were calculated. B) Correlation plot for depleted blood samples shown in A (uniquely mapping reads). Major globins (marked in red) are retained in samples depleted with HMR V2 and efficiently removed by RiboCop HMR+Globin depletion.
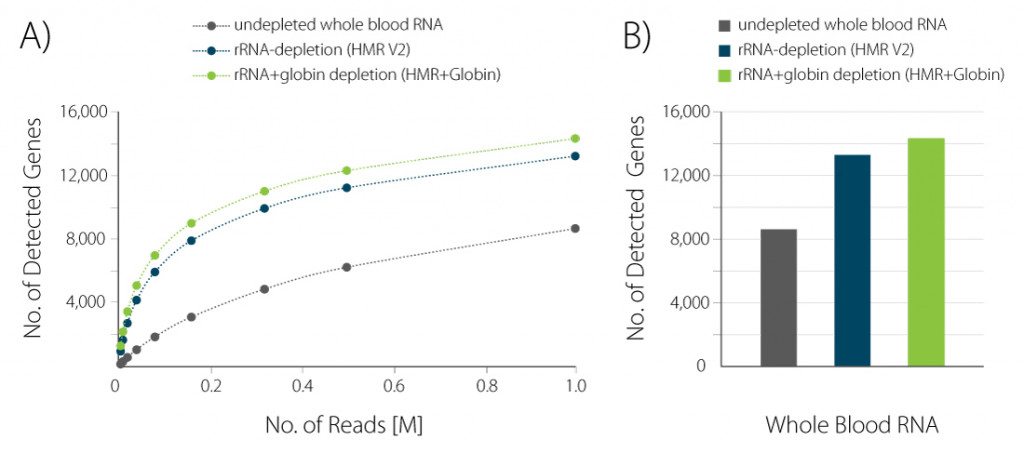
Figure 2 | Increased gene detection upon globin depletion. A) The number of detected genes per number of reads uniquely mapping to exons (FeatureCounts) was plotted for the samples shown in Figure 1. B) Bar plot illustrating the number of detected genes for the samples shown in Figure 1 at 1 M reads /sample, normalized to Counts Per Million (CPM) at a threshold of CPM > 1.
RiboCop Performance Across Species
RiboCop enriches RNAs of interest by efficiently removing rRNA and globin mRNA from human, mouse, and rat whole blood RNA (Fig. 3).
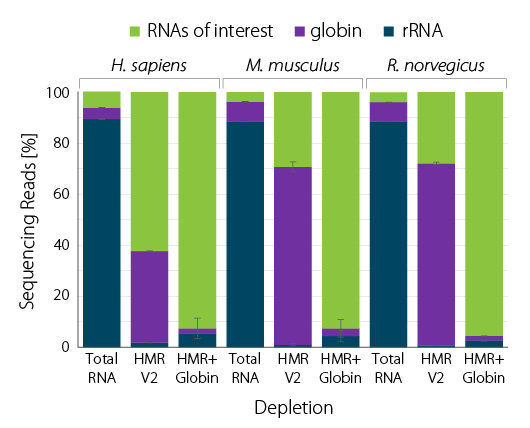
Figure 3 | RiboCop rRNA Depletion for Human/Mouse/ Rat plus Globin (HMR+Globin) enriches for RNAs of interest in blood sample across species. 100 ng whole blood RNA were depleted with either RiboCop HMR V2 or RiboCop HMR+Globin. Library preparation, sequencing, and data analysis was performed as described in Figure 1. Reads were mapped against the respective reference genomes for human (GRCh38.95), mouse (mmu_GRCm38.95), and rat (rno_Rnor_6.0.95), respectively.
Workflow
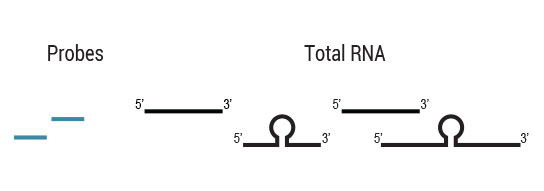
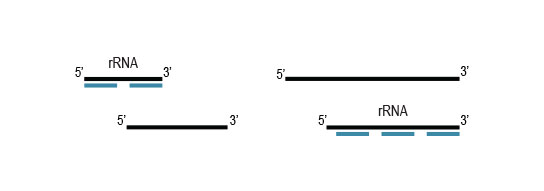
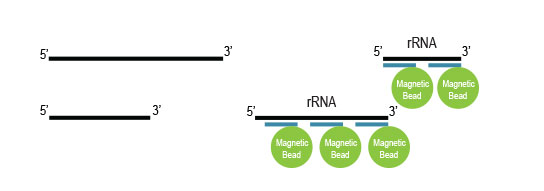
Samples are treated using a set of affinity probes for specific depletion of rRNA sequences. RiboCop probes efficiently remove rRNA and therefore afford a comprehensive view of transcriptome composition.
1 hr 30 min
30 min
Testimonial
“We have evaluated a double depletion method along with poly(A) capture using the CORALL RNA-Seq kit on isolated RNA from human blood collected in PAXgene tubes. When comparing combined rRNA and haemoglobin depletion (RiboCop rRNA Depletion Kit HMR plus Globin) with only using poly(A) capture (Poly(A) RNA selection kit), we saw less multimapping of reads. In addition, we managed to identify additional expressed genes whilst seeing a strong correlation between expression levels in genes identified in both sets. We were very pleased with this improvement. The workflow of using poly(A) selection combined with RiboCop HMR plus Globin and CORALL RNA-Seq library preparation was smooth and user-friendly and is highly recommended from a lab perspective. “
Elina Ekedahl, NGS Specialist, TATAA Biocenter
FAQ
Frequently Asked Questions
Access our frequently asked question (FAQ) resources via the buttons below.
Please also check our General Guidelines and FAQ resources!
How do you like the new online FAQ resource? Please share your feedback with us!
Downloads
Safety Data Sheet
If you need more information about our products, please contact us through support@lexogen.com or directly under +43 1 345 1212-41.
First time user of RiboCop?
First Time User? We’re excited to offer you an exclusive introductory offer.
Buy from our Webstore
Need a web quote?
You can generate a web quote by Register or Login to your account. In the account settings please fill in your billing and shipping address. Add products to your cart, view cart and click the “Generate Quote” button. A quote in PDF format will be generated and ready to download. You can use this PDF document to place an order by sending it directly to sales@lexogen.com.
Web quoting is not available for countries served by our distributors. Please contact your local distributor for a quote.