RiboCop rRNA Depletion Kits for Bacteria
- Depletion of 23S, 16S, and 5S rRNA
- Optimized Probe Mixes for monocultures or mixed communities
- Innovative probe design minimizing off-target effects
- 1 ng to 1 µg total RNA input (FFPE compatible)
RiboCop for Bacteria
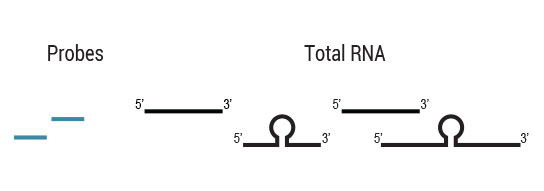
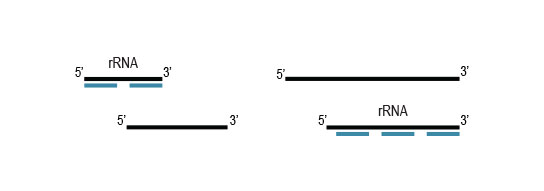
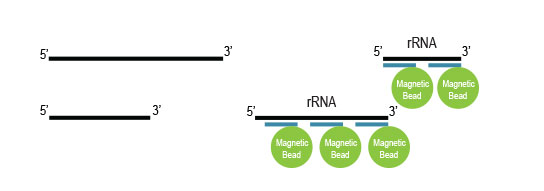
RNA extracted from bacterial species comprises up to 98 % of the ribosomal RNAs (rRNA), presenting a unique challenge especially when analyzing the transcriptome capacity from complex bacterial communities. Lexogen’s RiboCop rRNA Depletion Kits for Bacteria efficiently remove 23S, 16S, and 5S rRNA from mixed bacterial samples and monocultures.
RiboCop is offered as a stand-alone kit with the option to choose from three optimized Probe Mixes for depletion of rRNA from:
- Mixed bacterial samples (META, Cat. No. 125)
- Gram negative bacteria (G-, Cat. No. 126)
- Gram positive bacteria (G+, Cat. No. 127)
RiboCop is compatible with all random primed library prep kits such as the CORALL RNA-Seq V2 (Cat. No. 171 – 176).
Performance
Sequence what matters most! RiboCop enables specific, highly effective, non-enzymatic depletion of ribosomal RNA from a broad range of sample types.
Robust Performance Over a Wide Range of Input Amounts while Maintaining Unbiased Transcript Abundance
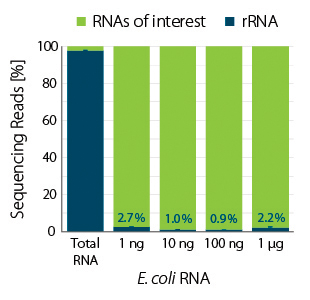
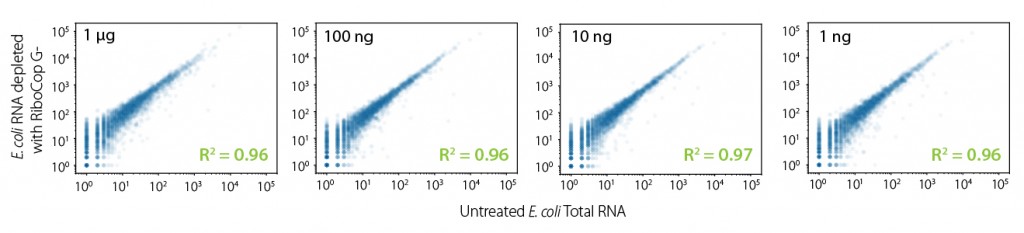
Ribosomal RNA was depleted from E. coli total RNA using RiboCop for Gram Negative Bacteria over a wide range of input amounts (1 ng to 1 µg). RiboCop for Bacteria efficiently reduces rRNA reads from 98 % to 1-3 % over a broad range of input amounts (Fig. 1) while maintaining unbiased transcript abundance (Fig. 2).
Figure 1 | RiboCop rRNA Depletion for Bacteria efficiently removes rRNA across a wide range of input amounts. NGS libraries were prepared using Lexogen’s CORALL Total RNA-Seq Library Prep Kit. Successful depletion was monitored by sequencing and subsequent analysis of remaining rRNA reads from untreated (Total RNA) and depleted E. coli RNA (1, 10, 100 ng and 1 µg). Sequencing on Illumina NextSeq (1×75 bp) and analysis by mapping to the MG1655 reference using BBMap. The percentage of reads mapping to rRNA is plotted in blue.
Figure 2 | RiboCop maintains unbiased expression profiles while efficiently removing undesired ribosomal RNA. Correlation of transcript abundance in untreated samples vs. samples depleted with RiboCop for Gram Negative Bacteria for 1 ng – 1 µg E. coli RNA input. Libraries were prepared using Lexogen’s CORALL Total RNA-Seq Library Prep Kit and sequenced on Illumina NextSeq (1×75 bp). Reads were mapped against the MG1655 reference genome using STAR aligner and counted with FeatureCounts (including multi mappers).
Performance on Meta-transcriptome Samples
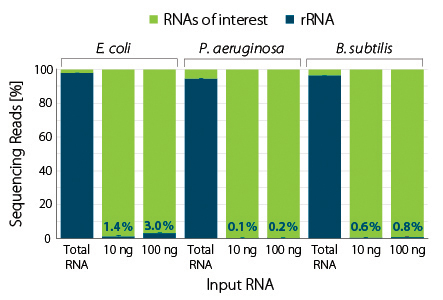
The RiboCop rRNA Depletion Kit for Mixed Bacterial Samples (META) is specifically designed for depletion of complex, mixed populations such as environmental communities or microbiome samples. Low quality microbiome samples from stool extractions were used for characterization of the Probe Mix designed for meta-transcriptomics (Tab. 1). The META Probe Mix can also be used for efficient and robust depletion of monocultures up to 100 ng total RNA input (Fig. 3) while maintaining unbiased transcript expression (Fig. 4).

Table 1 | Depletion rates for meta-transcriptome analyses. Different amounts of stool microbiome RNA was treated with RiboCop for Mixed Bacterial Samples (META). NGS-Libraries were prepared, sequenced, and analyzed as described in Fig. 1. Reads were mapped to ~60 annotations.
Figure 3 | RiboCop rRNA Depletion for Mixed Bacterial Samples (META) efficiently removes rRNA from various bacterial species. Two RNA amounts from monocultures of the indicated species were subjected to rRNA depletion using the META Probe Mix. Library preparation and sequencing were performed as described in Fig. 1. Reads were mapped against the respective genomes of E. coli MG1655, P. aeruginosa PAO1, and B. subtilis 168. The percentage of reads mapping to rRNA is plotted in blue.
Figure 4 | RiboCop META maintains constant transcript abundance across various species. Correlation of transcript abundance in untreated samples vs. samples depleted with RiboCop for Mixed Bacterial Samples (META) for 100 ng RNA input of three different species. Library preparation, sequencing, and data analysis were performed as described in Fig. 2. Reads were mapped against the E. coli MG1655, P. aeruginosa PA01, or B. subtilis 168 reference genomes, respectively.
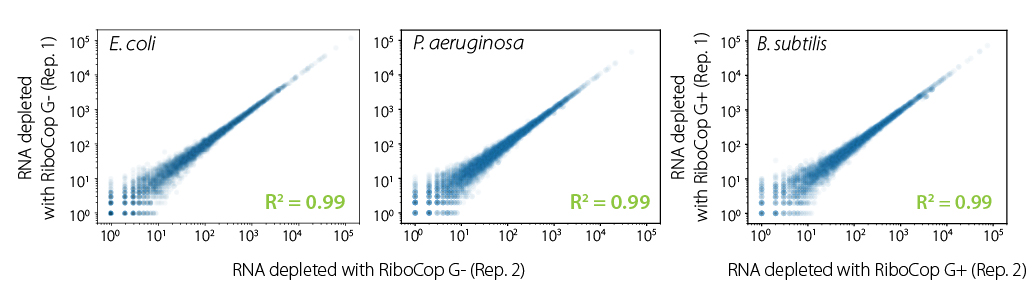
Excellent Reproducibility and Depletion Without Off-target Effects
RiboCop protocols are robust and highly reproducible. Correlation plots show excellent reproducibility between replicates (Fig. 5) independent of the species or the Probe Mix used for depletion.
Figures 2 and 4 additionally show a high correlation between transcript abundance in untreated vs. ribo-depleted samples for a wide input range (1 ng to 1 µg, Fig. 2) as well as across various species (Fig. 4) without outliers. This indicates that depletion with RiboCop for Bacteria occurs without of off-target effects.
Figure 5 | Excellent reproducibility between replicates. Correlation of replicates for two independent depletion reactions using 10 ng total RNA of the indicated species. Total RNA was ribo-depleted with RiboCop for Gram Negative or Gram Positive Bacteria, respectively. Library preparation, sequencing, and data analysis were performed as described in Fig. 4.
Workflow
Samples are treated using a set of affinity probes for specific depletion of rRNA sequences. RiboCop probes efficiently remove rRNA and therefore afford a comprehensive view of transcriptome composition.
1 hr 30 min
30 min
Testimonial
“We have extensively tested Lexogen’s RiboCop META rRNA depletion kit on bacteria for transcriptomics. Due to a research focus in infection biology at our university, we are working on RNA sequencing projects with various types of bacteria. The RiboCop META is a versatile tool with very good depletion results for all types of bacteria tested so far. Therefore, we would like to continue to use it for future transcriptome profiling projects.”
Tobias Heckel, Head Core Unit Systems Medicine (NGS & Bioinformatics), University of Würzburg, Germany
RiboCop for Bacteria Selection Tool

If you are uncertain which kit to choose, then the RiboCop for Bacteria Kit Selection Tool can help to find the best-suited kit for your species of interest.
FAQ
Frequently Asked Questions
Access our frequently asked question (FAQ) resources via the buttons below.
Please also check our General Guidelines and FAQ resources!
How do you like the new online FAQ resource? Please share your feedback with us!
Downloads
Safety Data Sheet
If you need more information about our products, please contact us through support@lexogen.com or directly under +43 1 345 1212-41.
First time user of RiboCop?
First Time User? We’re excited to offer you an exclusive introductory offer.
Buy from our Webstore
Need a web quote?
You can generate a web quote by Register or Login to your account. In the account settings please fill in your billing and shipping address. Add products to your cart, view cart and click the “Generate Quote” button. A quote in PDF format will be generated and ready to download. You can use this PDF document to place an order by sending it directly to sales@lexogen.com.
Web quoting is not available for countries served by our distributors. Please contact your local distributor for a quote.