Dear Customers,
SENSE mRNA-Seq Library Prep Kits (Cat. No. 001.24 and 001.96) are being discontinued. We recommend instead to use our CORALL mRNA-Seq Library Prep Kit.
Description
SENSE mRNA-Seq Library Prep Kit
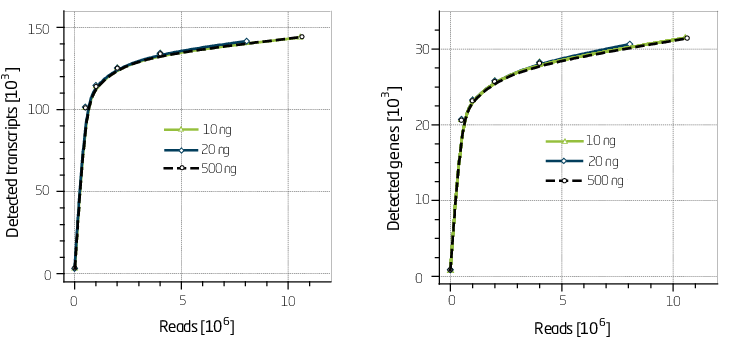
SENSE is a complete strand-specific mRNA-Seq library prep kit for accurate gene expression profiling, transcriptome sequencing, discovery and quantification of antisense transcripts and overlapping genes.
Superior Strand-Specificity
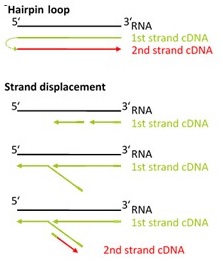
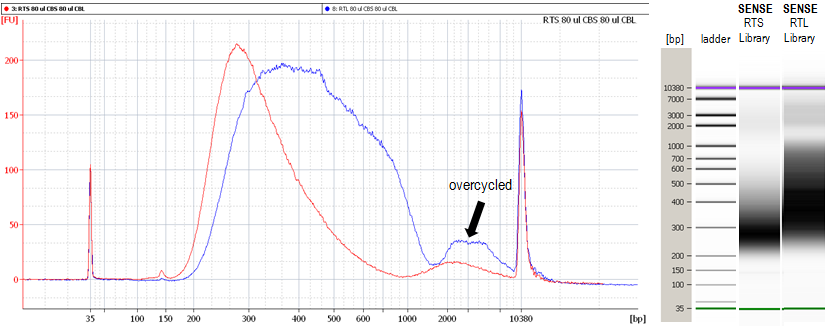
The strand-displacement stop/ligation technology used in SENSE generates fewer antisense artifacts which can be produced by template-switching in protocols which utilize RNA or cDNA fragmentation. This results in exceptional (>99.9 %) strand-specificity and reduced experimental noise, enabling the detection and quantification of antisense transcripts with high confidence.
Rapid Turnaround
NGS-ready libraries can be produced from total RNA samples in under 5 hours with less than 50 % hands-on time, allowing RNA extraction, library preparation and quality control to be performed in one day.
All-in-One Solution
No additional kits or reagents for poly(A) RNA selection, library amplification, size selection or purification, or barcodes are required.
Low Amount of Input Total RNA
The typical amount of input total RNA is 1 ng – 2 µg.
Efficient rRNA Elimination
Different Sequencing Read Length
For good representation and even coverage of all transcripts in your experiment the library should have a size suitable for the chosen sequencing read length. The size of SENSE libraries for Illumina can be adjusted by simply modulating appropriate buffers during reverse transcription/ligation and purification steps.
Compatibility
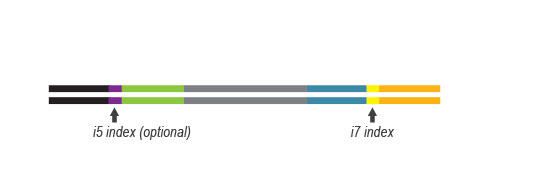
Simple Multiplexing
Workflow
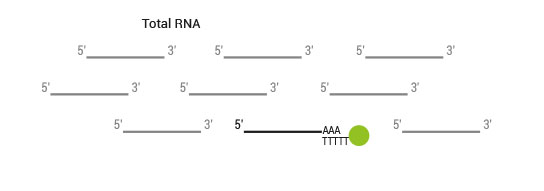
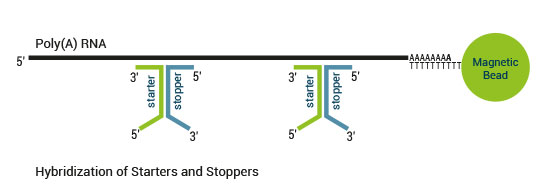
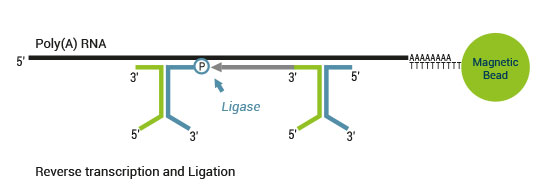
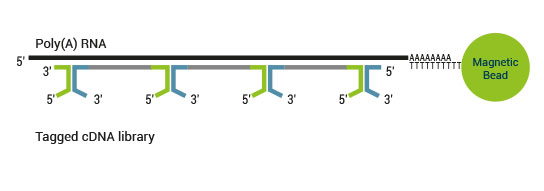
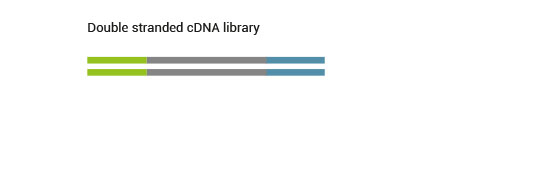
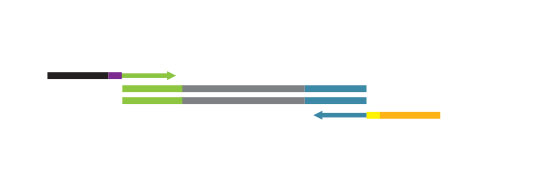
The SENSE kit features an all-in-one mRNA-Seq library preparation protocol. Most of the procedure is performed on magnetic beads, making it amenable to automation and decreasing purification time. The SENSE protocol has a simple workflow consisting of just 3 major steps.
Please find a schematic overview of the workflow for SENSE mRNA-Seq for Illumina below:
Library quantification can be performed with standard protocols, e.g., microcapillary electrophoresis or qPCR. Produced libraries are compatible with single-read or paired-end sequencing reagents.
Featured Publications
List of the most recent SENSE mRNA-Seq for Illumina Publications.
List of the most recent SENSE mRNA-Seq for Ion Torrent Publications.
Automation
autoSENSE mRNA-Seq V2 for Illumina
autoSENSE V2 is the automated solution of the SENSE mRNA-Seq Library Prep Kit V2 for Illumina in combination with the software and the Automation Module, which is currently available for the Perkin Elmer Sciclone NGS Workstation in combination with the Zephyr Workstation for the post-PCR step. Please contact info@lexogen.com if you are interested in automating the SENSE protocol on any other liquid handler.
The main advantage of automating the RNA-Seq library preparation process is in reducing sample tracking errors while dramatically increasing throughput.
Fast and Fully Walk-Away Protocol
The whole autoSENSE V2 workflow is a complete walk-away protocol and 96 samples can be run within 9 hours, including about 2 hours of manual setup time. Since pre- and post-PCR can be run on separate machines the protocol time can be reduced by parallelizing the workflow.
Compatible with SENSE mRNA-Seq V2
The autoSENSE protocol is identical to the manual version of SENSE mRNA-Seq Library Prep Kit V2 and the software is downloadable from our website. With the Automation Module different library size selections can be performed and the workflow can be fully automated.
Flexibility of the Throughput
The autoSENSE kit is appropriate for preparing 9,216 barcoded libraries. The liquid handler program allows for processing of samples in multiples of 8 reactions (full columns of a 96-well plate). The reagents from a single kit can be distributed over several machine runs.
Avoiding Cross Contamination
Pre- and post PCR steps can be programmed on different machines to reduce the risk of cross-contamination of the pre-PCR samples by PCR products.
FAQ
Frequently Asked Questions
Please find a list of the most frequently asked questions below. If you cannot find the answer to your question here or want to know more about our products, please contact support@lexogen.com.
Downloads
SENSE mRNA-Seq Library Prep Kit V2 for Illumina
![]() User Guide for Illumina – update 17.07.2019
User Guide for Illumina – update 17.07.2019
![]() PCR Add-on Kit for Illumina User Guide – update 03.01.2023
PCR Add-on Kit for Illumina User Guide – update 03.01.2023
![]() Lexogen i5 6 nt Dual Indexing Add-on Kits (5001-5096) User Guide – update 03.01.2023
Lexogen i5 6 nt Dual Indexing Add-on Kits (5001-5096) User Guide – update 03.01.2023
![]() SENSE Application Note
SENSE Application Note
![]() Lexogen i7 and i5 Index Sequences – update 05.05.2020
Lexogen i7 and i5 Index Sequences – update 05.05.2020
Material Safety Datasheets
MSDS Information can be found in the Documents page.
If you need more information about our products, please contact us through support@lexogen.com or directly under +43 1 345 1212-41.
SENSE Bioinformatics Data Analysis
Find more about the SENSE Data Analysis here.
Buy from our webstore
Need a web quote?
You can generate a web quote by Register or Login to your account. In the account settings please fill in your billing and shipping address. Add products to your cart, view cart and click the “Generate Quote” button. A quote in PDF format will be generated and ready to download. You can use this PDF document to place an order by sending it directly to sales@lexogen.com.
Web quoting is not available for countries served by our distributors. Please contact your local distributor for a quote.