GET THE BEST SEQUENCING RESULTS
with all-in-one library preparation kits
→ Our new CORALL RNA-Seq V2 Library Prep Kits
Do you have any questions?
Find the ideal kit for your application:
CORALL mRNA-Seq V2
Whole transcriptome mRNA sequencing is perfectly suited for expression analysis and delivers gene and transcript level quantification even for low input samples.
Get the best sequencing data with CORALL mRNA-Seq V2. The kit enables fast and cost-efficient generation of stranded, UMI labelled, and unique dual indexed libraries with adjustable insert size.
Performance
Exceptional Gene Detection Across a Wide Range of Input Amounts
CORALL mRNA-Seq V2 delivers excellent gene discovery rates across a wide range of RNA input amounts for long and medium library lengths (Fig. 1).
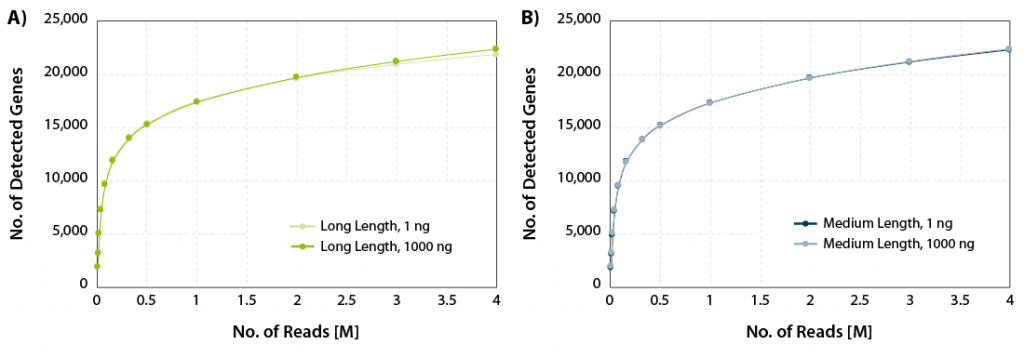
Figure 1 | Gene discovery rates. A) Gene discovery rates for libraries with long length (average size 550 bp) and B) medium length (average size 350 bp). 1 ng, and 1000 ng Universal Human Reference RNA (UHRR) were used as input for poly(A) selection and library preparation using CORALL mRNA-Seq V2. Libraries were sequenced on Illumina® NextSeq500 (2×150 bp). The number of detected genes is plotted against the number of reads mapping uniquely to exons (calculated with featureCounts).
Excellent Reproducibility and Sensitivity
Correlation analysis of CORALL mRNA-Seq libraries from 1 ng total RNA input prior to poly(A) selection reveals excellent reproducibility for libraries of all sizes (Fig. 2).
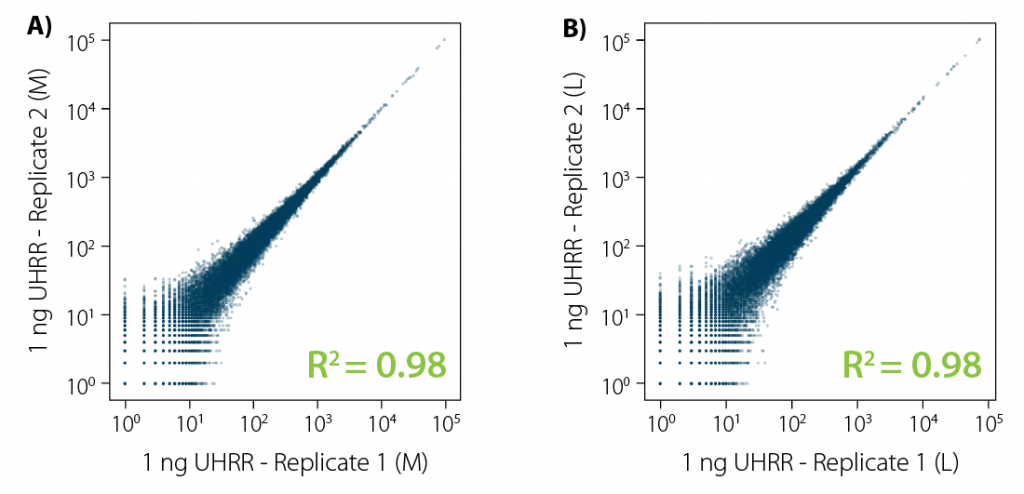
Figure 2 | Excellent reproducibility between replicates for low input RNA. Correlation analysis between replicates for CORALL mRNA-Seq with 1 ng UHRR input for libraries with A) medium length (average size ~350 bp) and B) long length (average size ~550 bp). Libraries were sequenced on Illumina® NextSeq500 (2×150 bp).
Excellent Isoform Detection Across a Wide Range of Input Amount
Lexogen’s Spike-in RNA Variant controls (SIRVs) are a set of 69 synthetic RNA molecules that mimic transcript isoform complexity. CORALL mRNA-Seq V2 authentically reproduces the expected coverage of the highly complex SIRV6 locus with 16 transcript isoforms and 2 anti-sense transcripts (Fig. 3). Thus, CORALL is ideal for isoform discovery and alternative splicing applications, even at inputs as low as 1 ng of total RNA (prior to poly(A) selection).
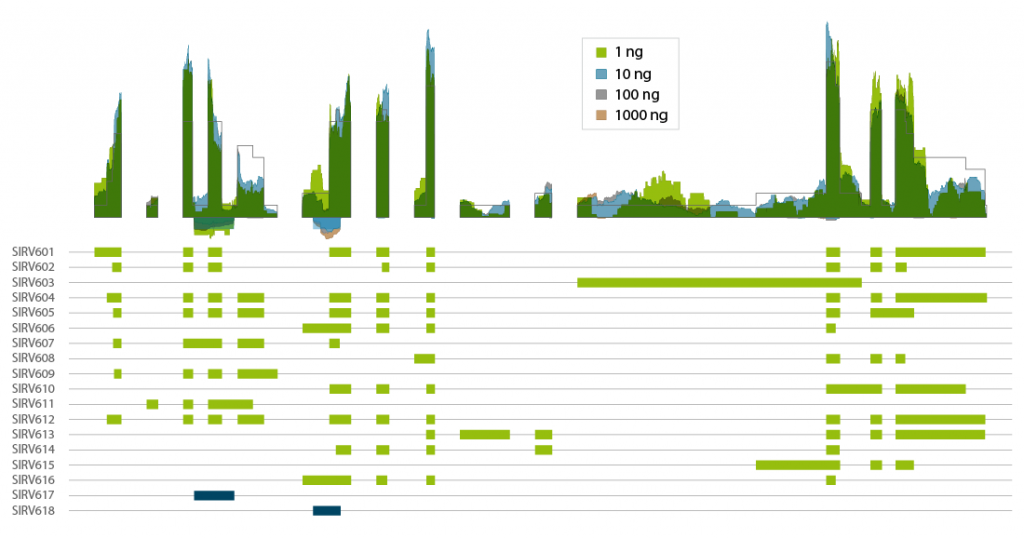
Figure 3 | Coverage transcript isoforms of SIRV6 across RNA input amounts. Lexogen’s SIRV-Set 3 was spiked into Universal Human Reference RNA at of 1 % of the mRNA fraction. Total RNA amounts of 1000 ng, 100 ng, 10 ng, and 1 ng were used for poly(A) selection and CORALL mRNA-Seq V2 library preps for generation of libraries with long insert sizes. Libraries were sequenced on Illumina® NextSeq500 (2×150 bp). Reads were mapped to the SIRV reference genome and visualized in condensed coverage profiles on gene level (upper panel). The exon-intron structure of all 18 transcripts of the SIRV6 locus is shown in the lower panel (with antisense transcripts in blue). The coverage generated by CORALL mRNA-Seq V2 is shown as overlay for four different input amounts.
Performance on Blood Samples
Total RNA from mammalian blood can be obtained by minimally invasive sampling and is therefore the most commonly used sample to study diseases in cohorts or patients. However, whole blood RNA is comprised of highly abundant undesired RNA species, such as ribosomal RNA (rRNA), accounting for ~80 – 90 % of total RNA, and globin mRNA, representing 30 – 80 % of all mRNAs.
As a consequence, the majority of reads in mRNA-Seq experiments from blood samples maps to globin (Fig. 4). While globin can be removed from fresh blood samples during the RNA extraction step, this process is not efficient when blood samples need to be stored of frozen prior to processing.
The combination of CORALL mRNA-Seq with an additional RiboCop depletion step for removal of rRNA and globin mRNA provides a convenient workflow to process also stored blood samples. This workflow frees up sequencing space for mRNAs of interest (Fig. 4) and leads to a significant increase in gene detection (Fig 5 A and B).

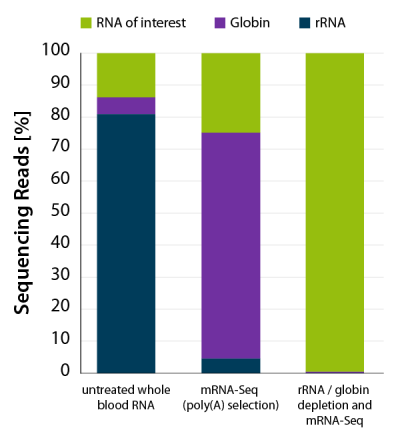
Figure 4 | CORALL mRNA-Seq V2 with an additional rRNA and globin depletion step efficiently removes globin mRNA from human whole blood RNA for mRNA-Seq applications. RNA was extracted from human donor whole blood. Whole blood RNA was enriched for polyadenylated transcripts using the Poly(A) Selection Kit included in the CORALL mRNA-Seq V2 bundles. One set of samples was additionally depleted with RiboCop HMR+Globin. Libraries were prepared with the CORALL RNA-Seq V2 Library Prep and sequenced on NextSeq500 (1×75 bp). Reads were mapped against the human reference genome (GRCh38.95) and reads mapping to rRNA (blue) and globin (purple) were calculated.
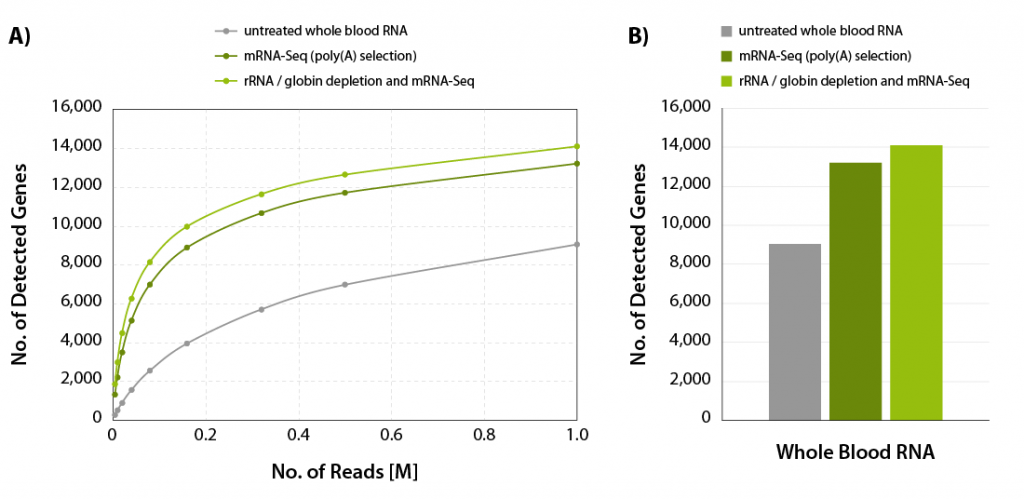
Figure 5 | Increased gene detection for CORALL mRNA-Seq with additional rRNA and globin depletion. A) The number of detected genes per number of reads uniquely mapping to exons (FeatureCounts) was plotted for the samples shown in Figure 4. B) Bar plot illustrating the number of detected genes for the samples shown in Figure 4 at 1 M reads /sample, normalized to Counts Per Million (CPM) at a threshold of CPM > 1.
Superior End-to-End Coverage
The innovative workflow of all CORALL versions delivers superior transcript start and end site coverage and exact representation transcription start sites.
Workflow
1 ng – 1 µg total RNA can be used for poly(A) selection. Poly(A) RNA is enriched by hybridization to oligo(dT) magnetic beads. Any RNA without poly(A) stretches is washed away and poly(A) RNA is finally eluted.
After elution, poly(A) RNA can directly be used as template for CORALL reverse transcription. No prior RNA fragmentation is necessary, insert size is determined during reverse transcription.
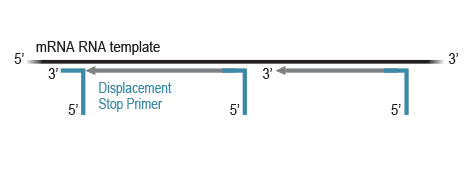
CORALL library generation is initiated by random hybridization of Displacement Stop Primers (DSP) with partial Illumina-compatible P7 sequences, to the RNA template. Reverse transcription extends each DSP to the next one, where transcription is effectively stopped. This stop prevents spurious second strand synthesis, maintaining excellent strand specificity.
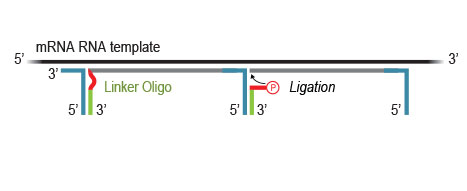
Highly efficient ligation of Linker Oligos to the 3’ ends of first-strand cDNA fragments then introduces partial Illumina compatible P5 sequences and Unique Molecular Identifiers (UMIs).
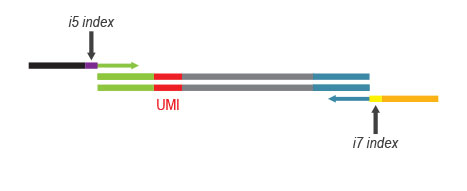
During PCR, second strand synthesis is performed, and the double-stranded cDNA is amplified. In doing so, unique dual i7 and i5 indices as well as complete adapter sequences required for cluster generation on Illumina instruments are added.
FAQ
Frequently Asked Questions
Access our frequently asked question (FAQ) resources via the buttons below.
Please also check our General Guidelines and FAQ resources!
How do you like the new online FAQ resource? Please share your feedback with us!
Downloads
CORALL mRNA-Seq Library Prep Kit
Safety Data Sheet
If you need more information about our products, please contact us through support@lexogen.com or directly under +43 1 345 1212-41.
First time user of CORALL V2?
First Time User? We’re excited to offer you an exclusive introductory offer.
Buy from our Webstore

Not sure which CORALL RNA-Seq product is right for you?
Try the CORALL Configurator and find the ideal kit for your application
in less than 2 minutes!
Need a web quote?
You can generate a web quote by Register or Login to your account. In the account settings please fill in your billing and shipping address. Add products to your cart, view cart and click the “Generate Quote” button. A quote in PDF format will be generated and ready to download. You can use this PDF document to place an order by sending it directly to sales@lexogen.com.
Web quoting is not available for countries served by our distributors. Please contact your local distributor for a quote.