
GET THE BEST SEQUENCING RESULTS
with all-in-one library preparation kits
→ Our new CORALL RNA-Seq V2 Library Prep Kits
Do you have any questions?
Find the ideal kit for your application:
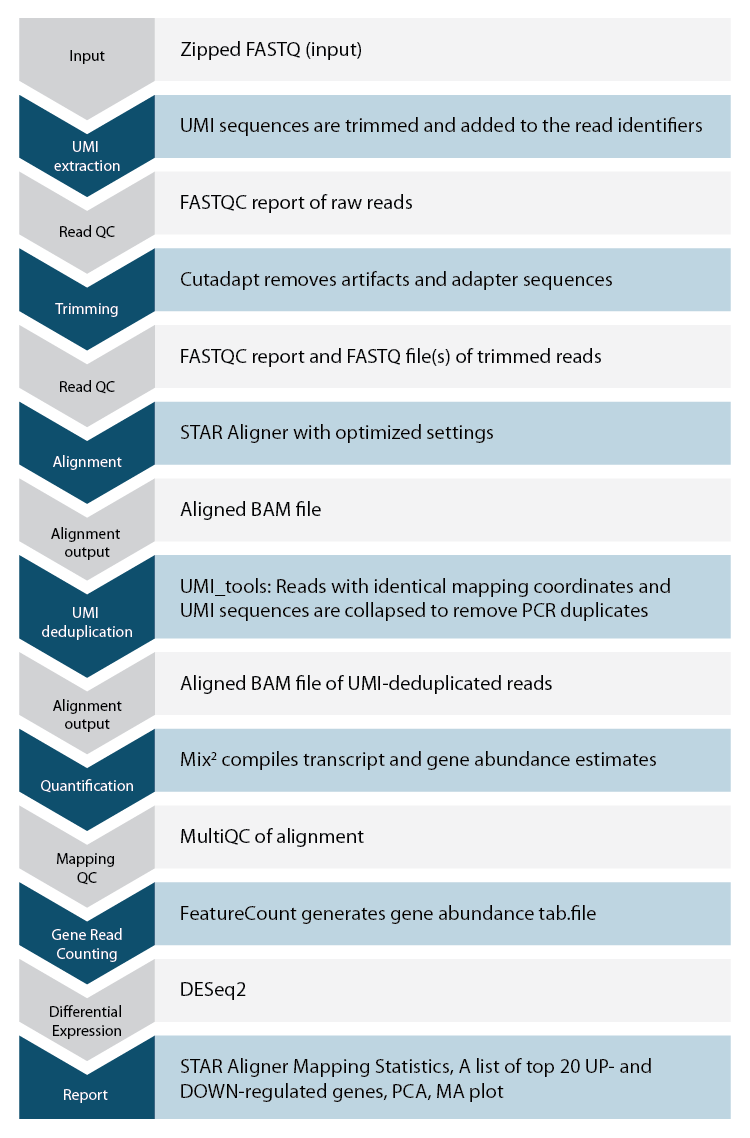
Lexogen’s Automated Data Analysis Solution
Lexogen offers an automated solution for CORALL data analysis based on our state-of-the-art proprietary pipeline, which allows researchers to analyze CORALL samples in a convenient and fast way. The workflow for CORALL data analysis pipeline is shown on Figure 1.
Codes for CORALL data analysis on Lexogen’s automated data analysis solution are available for purchase. Please contact us at sales@lexogen.com.
Sequence of Indices (Barcodes) Used for Multiplexing
CORALL libraries can be easily multiplexed. External barcodes allowing up to 9,216 samples to be uniquely indexed are available for multiplexing. Up to 96 i7 indices (i7 Index Plate) are included in the CORALL kit and additionally 96 external i5 indices are available in the Lexogen i5 6 nt Dual Indexing Add-on Kits (Cat. No. 047).
This section describes a basic bioinformatics workflow for the analysis of CORALL NGS data and is kept as general as possible for integration with your standard pipeline. For more information please contact info@lexogen.com.
In contrast to most other library preparation protocols, CORALL libraries generate reads in forward orientation, thus mapping should be performed to the corresponding strand of the genome.
Demultiplexing
Demultiplexing can be carried out by the standard Illumina pipeline. Lexogen’s i7 6 nt index sequences are available for download here.
Processing Raw Reads
We recommend the use of a general fastq quality control tool such as FastQC or NGS QC Toolkit to examine the quality of the sequencing run. These tools can also identify over-represented sequences, which may optionally be removed from the data set.
Trimming
As CORALL libraries are based on random priming the first 9 nucleotides of Read 2 may have an increased error rate. As random priming may also occur at the junction between the ultimate exon and the poly(A) tail, mapping rates can be increased by trimming of poly(A) sequences at the 3′ end of Read 1 and poly(T) sequences the 5′ end of Read 2, when analyzing data from paired end runs. Further, CORALL libraries contain N12 Unique Molecular Identifiers (UMIs) at the start of Read 1. Hence, the first 12 nucleotides of Read 1 can be trimmed before proceeding to alignment. Alternatively, a less stringent aligner could be used with relaxed settings. Low quality sequences and adapter sequences should be trimmed. In case an adapter sequence is detected at the 3’ end of Read 2, an additional 12 nucleotides upstream of the adapter can also be trimmed (i.e., the UMI sequence).
Alignment
After trimming, filtered and trimmed reads can be aligned with a short read aligner to the reference genome or assembled de novo. Please note, that Read 1 reflects the RNA transcript sequence not the cDNA sequence. This is important for downstream applications. If data from paired-end runs with read length >100 nucleotides is analyzed, ensure that the aligner used can handle overlaps (e.g., use relaxed settings).
Read Counting and Downstream Analyses
Depending on the intended application different methods for read counting on transcript or gene-level can be applied in order to generate expression data.
The analysis of SIRV spike-in control reads can be performed by aligning the trimmed reads to the SIRVome and evaluating the number and levels of detected isoforms. The SIRVome .fasta and .gtf annotation files are available for download from www.lexogen.com/sirvs/downloads.




