for Ultra-high-throughput Testing
- Results in 20 hours
- Scalable to population size screening
- High specificity of 99.8 %
- Saving scarce consumables
Lexogen’s SARS-CoV-2 Test
- The most economical solution for true mass screening
- Flexible: analyze 768 to 36,864 samples in a single Next Generation Sequencing (NGS) run
- Scalable to population size
- Get results within 20 hours
- Higher specificity for SARS-CoV-2 than antigen testing: 99.8 % for samples with Ct ≤30
- Based on Lexogen’s industry-leading QuantSeq and QuantSeq-Pool NGS technologies
- Save significant amounts of plastic ware compared to other NGS-based approaches
“Thanks to an initiative on the Vienna Biocenter Campus, our company is screening each employee every three days. This allows us to identify (and quarantine) infected individuals early enough to stop the spread of the virus. At this frequency, so far none of our infected employees have contaminated their colleagues. This actually helped us to finish the rapid development of this mass test.” Stéphane Barges, Chief Executive Officer, Lexogen GmbH
“Testing infected people regularly after infection, could allow them to return to work as soon as possible with confidence when they are not infectious anymore. This would save a tremendous amount of labor days and avoid a temporary closing of companies or of some activities.” Alexander Seitz, Chief Innovation Officer and Founder, Lexogen GmbH
Official Press Release in English
Offizielle Pressemitteilung in deutscher Sprache
Mass Screening
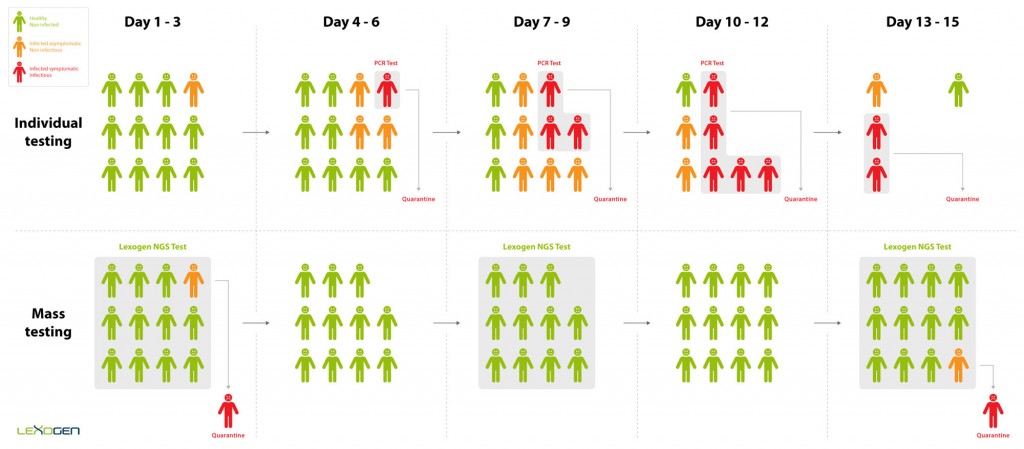
The impact of the Coronavirus pandemic has called for a new, more efficient screening approach to identify and isolate infected individuals. Current testing methods are limited in their throughput and do not allow for the scale of screening required to test repeatedly at population scale. Lexogen has therefore developed the QuantSeq-Pool Targeted SARS-CoV-2 Panel, a targeted RNA sequencing kit that leverages the ultra-high throughput capacity of NGS. Early sample barcoding allows to trace individual samples throughout the workflow while triple indexing enables the analysis of millions of samples in parallel. This way, the panel allows to quickly identify and isolate infected individuals even if they are asymptomatic. These individuals are then quarantined before they are infectious, thereby stopping the spread of the disease (Figure 1).
Results in 20 Hours
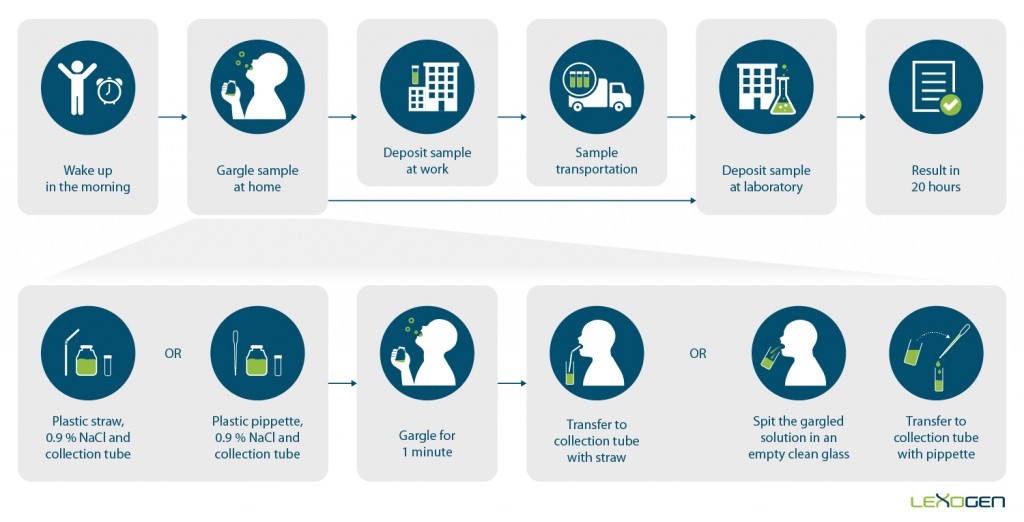
This new COVID test works with RNA extracted not only from nasopharyngeal but also from conveniently obtained gargle samples, making the procedure painless and allowing for self-sampling. Lexogen offers the SARS-CoV-2 Rapid RNA Extraction Kit for extraction of 96 samples in 15 minutes with full automation suitability for high-throughput processing (Figure 2).
Comparison of COVID-19 Test Methods
| Lexogen Panel (NGS, RNA-Seq) | PCR (RT-qPCR) | LAMP (RT-LAMP) | Antigen (Ag) | Benefit | |
|---|---|---|---|---|---|
| Target | Viral RNA | Viral RNA | Viral RNA | Viral antigen | |
| Method | Targeted conversion of RNA to DNA, library preparation and ultra high-throughput sequencing, bioinformatic data evaluation. | Targeted conversion of RNA to DNA and amplification of DNA which is visualised on a real-time fluorescence measurement. | Conversion of RNA to DNA and amplification of DNA which is visualised by a color change in the reaction tubes. | Antibody binds to a protein on the surface of the virus and is visualised on a lateral flow strip. | |
| Sputum sampling (gargling) |
|
|
| ✖️ |
|
| Nasal sampling (nasopharyngeal) |
|
|
|
|
|
| Testing at Point of Care (PoC) | ✖️ | ✖️ | ✖️ |
|
|
| Testing site and trained personnel required for sampling | No | No / Yes | No / Yes | Yes |
|
| Max number of samples per run | 36,864 | 96 | 96 | 1 |
|
| Pooling | of individual molecular-barcoded samples | of samples w/o retaining identity | of samples w/o retaining identity | N/A |
|
| Sensitivity for samples with Ct ≤ 30 | > 99% | 100% | 97.50% | 84% – 97.6% |
|
| Specificity | 99.80% | 98% – 100% | 99.70% | > 97% |
|
| Suitable for symptomatic individuals |
|
|
|
|
|
| Suitable for asymptomatic individuals (screening) |
|
|
| ✖️ |
|
| Lab / Assay time | 16 h | 2 h | 2 h | 15 min | |
| Sample-to-result time | 20 h | 5 h – 2 days | 6 h | 15 min |
|
| < € 12 | € 40 – € 60 | € 9 – € 12 | € 7.10 – € 13 | ||
| Additional technical costs | RNA extraction | RNA extraction | ✖️ | ||
| Total cost per sample | < € 15 | > € 110 | > € 15 | > € 20 | |
| Further costs | sample transport | sample transport | sample transport | Testing sites to be set up and sampling done by trained personnel with protective equipment | |
Recommendations for Companies and Institutions
To reduce the economic and social impact of the pandemic we suggest the following approach for every public and private company as well as for public institutions (especially schools and healthcare institutions):
- Perform a regular testing of all employees (to avoid leakage) every three days to identify and quarantine infected individuals early enough to avoid infection of K1 (contact cases).
- Perform a daily testing of infected people (after 4 or 5 days) to assess their level of infectivity and get them back to work as soon as they are no more infectious.
What are the consequences of these preventive measures?
- It becomes unnecessary to quarantine all close contact persons.
- Infected people who are not infectious anymore could be back to work before the end of the current long quarantine period without endangering their colleagues.
What are the benefits of these preventive measures?
- Reduce the death toll and number of infected.
- Create a safe working environment that also reduces the stress for the employees (as per our experience).
- Maintain schools, hospitals, and elderly homes open while providing a safe workspace and more serenity for all the stakeholders (pupils, parents, teachers, patients, healthcare workers, etc.).
- Avoid a lockdown and keep the economic activities up and running to ensure a continuous revenue stream for the companies and the country.
- Reduce the dramatic pandemic negative impact on the EBIT of companies and GDP of countries.
SARS-CoV-2 Rapid RNA Extraction Kit
In conjunction with the new QuantSeq-Pool Targeted SARS-CoV-2 Panel for mass testing of Covid-19, Lexogen has co-developed a fast and automatable protocol for extraction of nucleic acids from SARS-CoV-2 test samples.
- Extraction of SARS-CoV-2 RNA in only 15 min
- Based on magnetic beads, no phenol/chloroform extraction
- Fully Automatable for high throughput
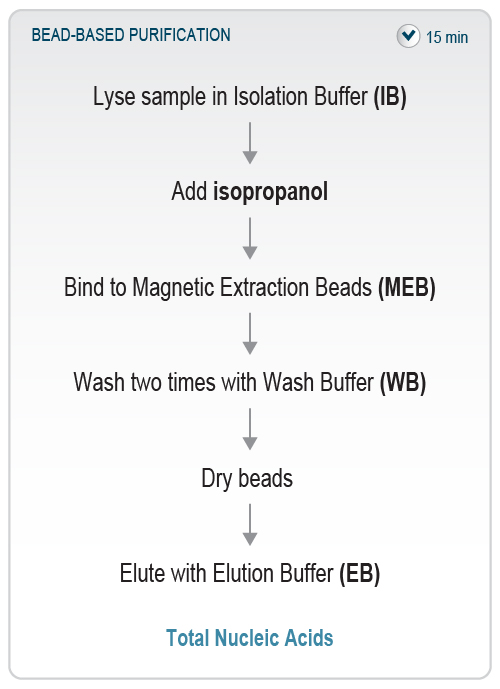
Figure 3 | Overview of the SARS-CoV-2 Rapid RNA Extraction workflow. The bead-based protocol extracts nucleic acids from the sample in only 15 minutes and is fully automatable.
The SARS-CoV-2 Rapid RNA Extraction Kit provides a streamlined protocol for isolation of nucleic acids from liquid samples such as nasopharyngeal and buccal swabs or gargle samples within 15 minutes and can be automated on liquid handlers. It is a quick and easy bead-based purification and can be directly used for molecular biology applications such as qRT-PCR, reverse transcription, and targeted RNA Sequencing.
First, the liquid sample is lysed in a highly chaotropic Isolation Buffer. The RNA is then bound onto Magnetic Extraction Beads by adding isopropanol to the lysed sample. After washing and drying of the beads, the bound nucleic acids are then eluted using pre-warmed Elution Buffer (Fig. 3). Isopropanol and ethanol for preparation of Wash Buffer have to be supplied by the user.
The resulting nucleic acids can be directly used for Lexogen’s QuantSeq-Pool Targeted SARS-CoV-2 Panel.
QuantSeq-Pool Targeted SARS-CoV-2 Panel
Lexogen has developed a new SARS-test, the QuantSeq-Pool Targeted SARS-CoV-2 Panel for Illumina. It is Next-Generation Sequencing based for scalability to ultra-high throughput and detects viral RNA extracted from conveniently obtained gargle samples as well as from nasopharyngeal swabs (and similar). Specificity for SARS-CoV-2 is very high (99.8 % for samples that have a Ct ≤30 in RT-PCR analyses).
- The most economical solution for SARS-CoV-2 mass screening: Batch processing reduces time requirements and early pooling saves significant amounts of plastic ware
- Analysis of up to 36,864 samples per NGS run, scalable to population size (millions of samples)
- Only 20 hours from sampling to results
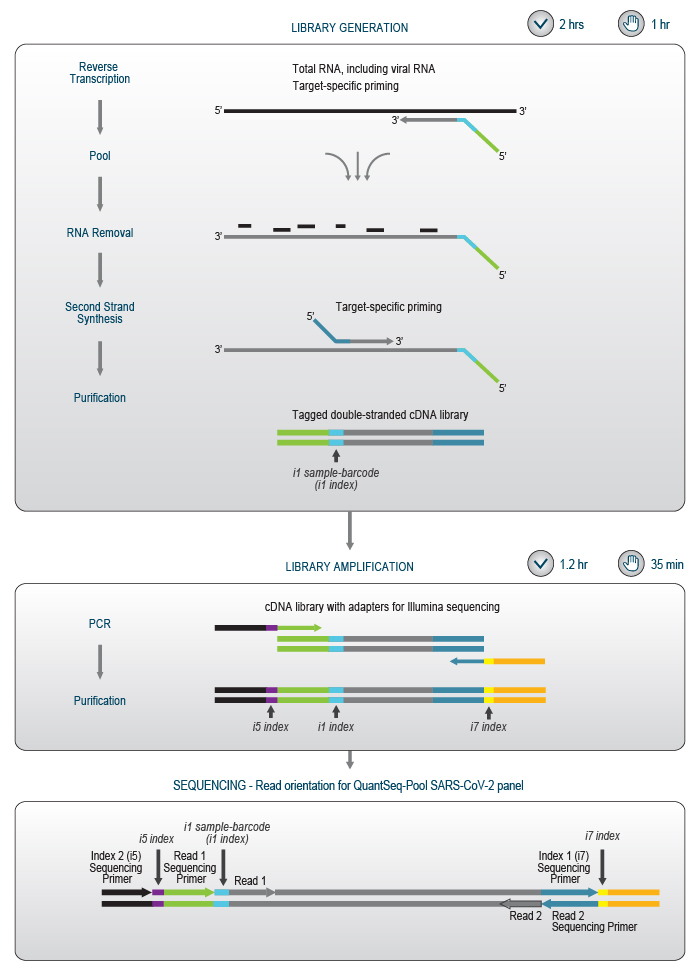
Figure 4 | Schematic overview of the QuantSeq-Pool SARS-CoV-2 Panel library preparation workflow. Read 1 corresponds to the cDNA sequence, i.e., the reverse complement of the target RNA sequence.
In the first step, extracted RNA is added to primers which are dried in 96-well plates for robust and easy handling. The primer plate also contains a synthetic positive control to validate functionality of the test for each individual sample. In the subsequent cDNA synthesis, four regions of the viral RNA genome are reverse transcribed, and samples are barcoded individually. The cDNAs of all samples are then pooled and transferred to a single well in another 96-well plate for second strand synthesis and PCR amplification (Fig. 4).
In the library amplification step, each cDNA pool obtains one of 384 Unique Dual Indices (UDIs), enabling multiplexing of up to 36,864 samples for analysis in a single NGS run. Early sample-barcoding allows to trace individual samples throughout the workflow while triple indexing enables the analysis of millions of samples. (Fig. 5).
Early pooling and batch processing significantly decrease overall and hands-on time and reduce the need for plasticware, not only driving down costs but also alleviating the pressure on supply chains in a highly volatile pandemic period. Massive savings are further leveraged by ultra-high multiplexing and the possibility to perform the NGS run economically in single-read mode with read lengths starting at 50 bp. The complete workflow is bead-based providing for automation and parallel preparations at lowest cost, and state-of-the-art indexing minimizes the risk of sample misassignment by using Lexogen’s patent-applied 12 nt UDIs and sample-barcodes.
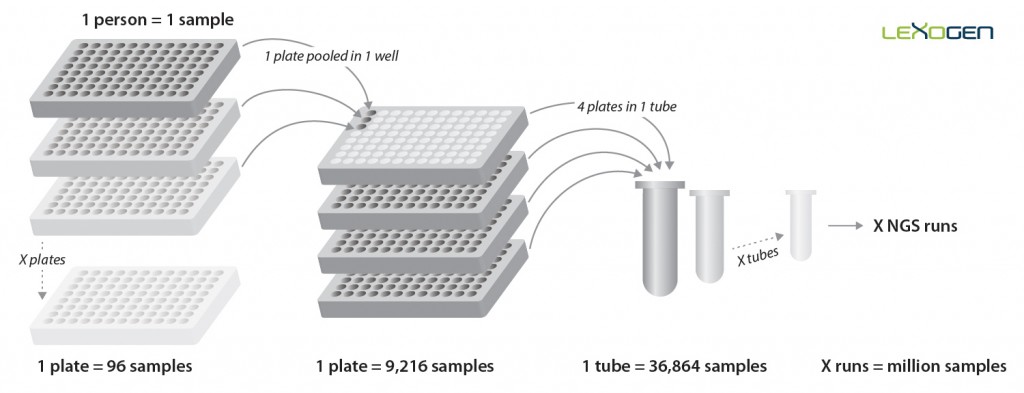
Figure 5 | Pooling strategy for the QuantSeq-Pool Targeted SARS-CoV-2 Panel. Each sample is barcoded during reverse transcription in a 96-well plate. These 96 cDNA samples are then pooled into a single well of another set of plates for PCR amplification which adds another layer of barcoding by introducing one of 384 UDIs. Thereby, up to 36,864 samples can be multiplexed in a single NGS run, allowing for the analysis of millions of samples in parallel setups.
The SARS panel has been extensively validated with RNA extracted using Lexogen’s SARS-CoV-2 Rapid RNA Extraction Kit, a fully automatable solution to obtain viral RNA in 15 minutes.
Contact & Kit Sizes
Contact
If you are interested in further information on the protocols, consulting on screening setups, and purchasing options please e-mail to covid-info@lexogen.com.
Products and Bundle
- 140.8×96 QuantSeq-Pool Targeted SARS-CoV-2 Panel for Illumina (768 reactions)
- 142.8×96 SARS-CoV-2 Rapid RNA Extraction Kit (768 preps)
- 164.8×96 QuantSeq-Pool Targeted SARS-CoV-2 Panel with Rapid RNA Extraction (bundle of Cat. No. 140 and Cat. No. 142, 768 reactions each)
Bulk sizes are available for large screening setups.