Blood is a highly informative, easily accessible, and minimally invasive sample type routinely used for diagnostic purposes and in clinical research to study pathological and physiological conditions and the impact of therapies. Whole blood transcriptome sequencing is an RNA sequencing (RNA-Seq)-based approach that provides in-depth gene expression profiling of the whole blood, giving insight into the expression and abundance of all transcripts on a system-wide scale. The development of high-throughput RNA-Seq technologies enables the collection of high-quality blood transcriptome information at a low cost, revolutionizing the discovery of biomarkers and our understanding of drug responses, pathological and physiological processes, and the non-invasive diagnosis of diseases.
The advantage of peripheral blood is that it is very easy to collect quickly, even from large cohorts of individuals. However, problems begin once blood is out of the body. In the past, researchers had to isolate white blood cells from whole blood before RNA isolation to deal with the plethora of contaminants in blood. Nowadays, protocols developed especially for preparing blood samples enable us to obtain high-quality RNA-Seq data from whole blood.
This blog article will discuss various challenges when performing RNA-Seq from blood samples and how to overcome them.
Collection of Blood
The first step in any blood transcriptome profiling experiment is the collection of blood, either by venipuncture or by a fingerstick (Rinchai et al., 2022). Advances in obtaining good-quality RNA from even small amounts of blood, such as when sampling is done via fingerstick, will enable extensive studies where participants could even perform self-sampling.
Once blood is out of the body, it is essential to prevent or reduce RNA degradation, which can occur during collection, storage, and processing (Rainen et al., 2002). One of the reasons why blood RNA is so prone to degradation is the high concentration of ribonucleases (RNases) in blood plasma (Bender et al., 2020). The power of RNases was demonstrated in a study where free RNA was incubated with blood plasma and degraded in only 15 seconds such that 99 % of the RNA could not be amplified via real-time PCR (Tsui et al., 2002). The best practice for preventing RNA degradation includes immediate and quick RNase deactivation upon blood collection.
To limit RNA degradation, and have ease of mind, especially when running large-scale blood transcriptomics projects, collecting the blood using commercially available tubes, such as PAXgene™ blood RNA tubes or Tempus™ blood RNA tubes (Rainen et al., 2002; Skogholt et al., 2017) is recommended. These vials contain proprietary reagents that stabilize RNA, yielding gene expression data that accurately reflect the blood’s state at sampling (Skogholt et al., 2017). However, substantial differences in gene expression profiles between the same blood samples collected in tubes from different providers (Menke et al., 2012; Nikula et al., 2013), suggest that the best practice is to use only one type of tube per experimental setting or if the combination of sampling systems is inevitable, always to run a control experiment and test for a common protocol (Skogholt et al., 2017).
Extraction of RNA from Blood and Genomic DNA Removal
Both PAXgene™ and Tempus™ tubes come with an (optional) RNA extraction protocol, which uses silica-membrane-based spin-columns to capture RNA. However, this is not a must as RNA stabilized in these tubes can be isolated using a different RNA extraction protocol, e.g., using acidic phenol / chloroform extraction, or some other protocol compatible with blood samples.
After the disruption of cellular walls, lysate will contain RNA, but also DNA and proteins. Genomic DNA is one of the “contaminants” that can linger throughout the entire RNA-Seq workflow and cause biases and quantification issues during the data analysis steps. Treatment with DNases to remove carry-over genomic DNA can be omitted in certain experimental setups, however, since blood cells contain more DNA than RNA, we strongly recommend performing DNase treatment on RNA isolated from blood, even when extraction was done using acidic phenol / chloroform. In addition, upon lysis of the cells, large amounts of DNA and proteins can lead to highly viscous samples, which are difficult to handle. To prevent this, we recommend reducing the starting material.
RNA Pre-treatment: Globin mRNA and rRNA Removal
Deciding if any RNA pre-treatment is needed before embarking on RNA-Seq library preparation heavily depends on experimental design, chosen library prep, and sample type. In RNA samples isolated from blood, two significant “contaminants”, occupying significant sequencing space, are globin mRNAs and ribosomal RNAs (rRNAs) (Mastrokolias et al., 2012; Krjutškov et al., 2016; Jang et al., 2020).
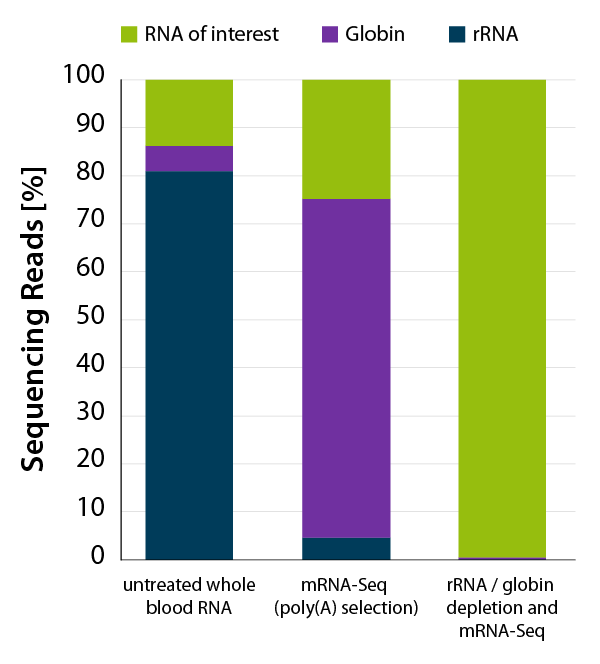
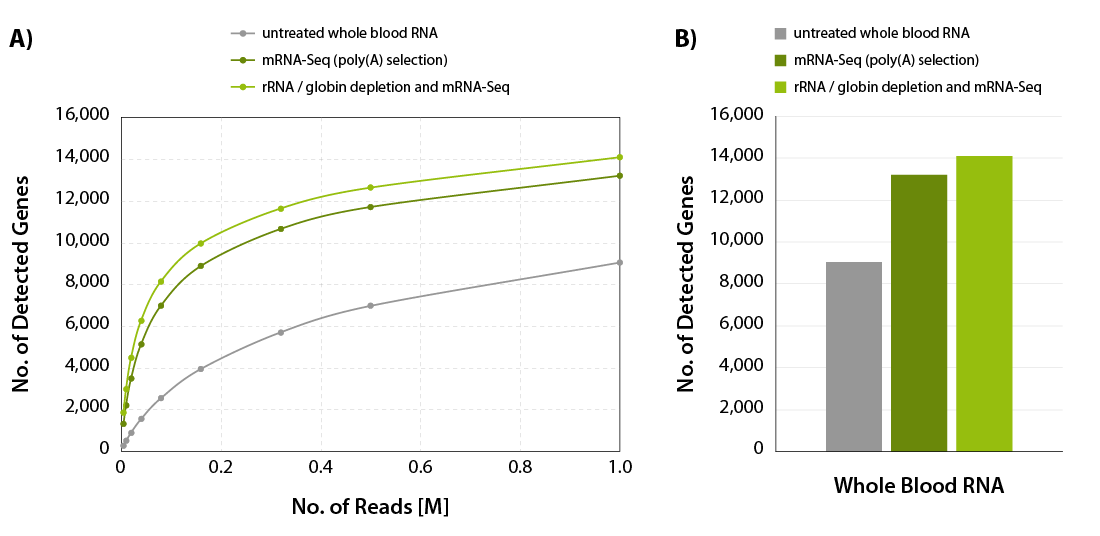
The amount of globin mRNA in red blood cells can account for 30-80 % of all mRNAs in the sample (Mastrokolias et al., 2012; Shin et al., 2014; Krjutškov et al., 2016). These transcripts consume a significant percentage of sequencing reads after poly(A) selection in an mRNA-Seq experiment (Figure 1, middle bar). Moreover, they also hamper the analysis of the transcriptional profile of blood (Shin et al., 2014; Krjutškov et al., 2016). In the absence of globin depletion, gene detection rates are reduced, as shown in Figure 2A and 2B.
Figure 1 | Lexogen’s CORALL mRNA-Seq V2 with rRNA and globin depletion efficiently removes globin mRNA from human whole blood RNA for mRNA-Seq applications. Whole blood RNA extracted from human donor was enriched for polyadenylated transcripts using the Poly(A) Selection Kit included in the CORALL mRNA-Seq V2 bundles. One set of samples was additionally depleted with RiboCop HMR+Globin. Libraries were prepared with the CORALL RNA-Seq V2 Library Prep and sequenced on NextSeq500 (1×75 bp). Reads were mapped against the human reference genome (GRCh38.95) and reads mapping to rRNA (blue) and globin (purple) were calculated.
Figure 2 | Increased gene detection for CORALL mRNA-Seq with rRNA and globin depletion. A) The number of detected genes per number of reads uniquely mapping to exons (FeatureCounts) was plotted for the samples shown in Figure 2. B) Bar plot illustrating the number of detected genes for the samples shown in Figure 2 at 1 M reads /sample, normalized to Counts Per Million (CPM) at a threshold of CPM > 1.
In addition to globin mRNAs, rRNAs are another undesired RNA species, accounting for 80-90 % of total RNA (Jang et al., 2020). rRNA depletion from RNA samples isolated from blood allows to free up additional valuable sequencing space and to focus sequencing reads to what matters the most (Figure 1).
We have developed a convenient workflow to process both fresh and stored blood samples for mRNA-Seq (Figure 3). The combination of CORALL mRNA-Seq V2 with RiboCop Human/Mouse/Rat + Globin, where both rRNAs and globin mRNAs are depleted provides a workflow that frees up sequencing space for mRNAs of interest (Figure 1), and it also leads to a significant increase in gene detection (Figure 2 A and B). This protocol focuses sequencing reads on mRNAs of the blood and allows the analysis of isoform abundance and splice variants, as well as polyadenylated non-coding RNAs.
Figure 3 | CORALL mRNA-Seq V2 protocol for whole blood, combined with an additional rRNA and globin depletion step, increases usable reads for mRNA-Seq applications.
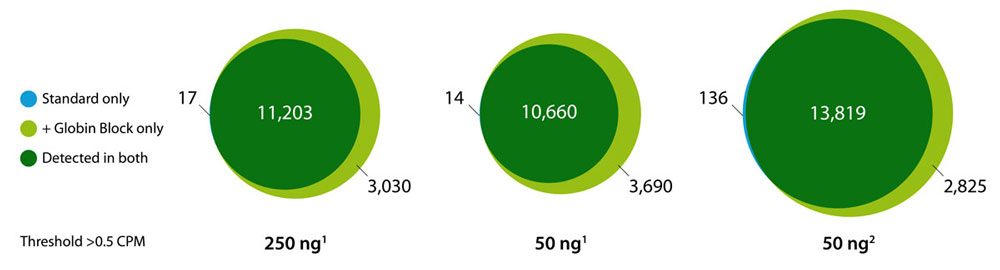
When the focus of research is to get insight into gene expression profiling only, 3’ mRNA-Seq is an excellent choice. 3’ mRNA-Seq, by design, allows skipping pre-processing steps such as poly(A) enrichment or rRNA depletion, however, globin depletion is highly recommended. When Lexogen’s QuantSeq 3’ mRNA-Seq library prep is combined with Globin Block (RS-Globin Block, Homo sapiens module), which prevents the generation of library fragments from globin mRNAs, gene detection rates increase dramatically, as shown in Figure 4.
Figure 4 | Increased gene detection in human blood QuantSeq libraries using Globin Block. CPM = Counts Per Million. Input RNA (50 ng, or 250 ng) was extracted with: 1 SPLIT RNA Extraction Kit, 2 other kit including red blood cell lysis.
Summary
Whole blood is a valuable, readily available, but also challenging sample type to prepare for RNA-Seq. Depending on the experimental design and type of RNA-Seq chosen, steps to acquire high-quality RNA-Seq data from whole blood will vary. No matter which approach is chosen, best practice advice is to prevent RNA degradation by inactivating RNases immediately upon sampling, treat RNA sample with DNase for gDNA removal, remove globin mRNAs for improved gene detection, and in case of mRNA-Seq or total RNA-Seq perform rRNA removal as well, to free up additional sequencing space.
References:
Written by Masa Ivin, PhD