Introduction
Drug discovery and development is an elaborate and lengthy process, and researchers are constantly seeking innovative tools to unravel the complexities of human biology further and accelerate the path to effective treatments. RNA sequencing (RNA-Seq) has emerged as a key method in drug discovery and development, and in this article, we will explore how RNA-Seq is revolutionizing drug discovery and development. Finally, we will introduce the benefits of incorporating temporal information into drug discovery research, and focus on technologies that can be used in combination with RNA-Seq to obtain information about RNA kinetics, which can be a game-changer in drug discovery.
What does a typical drug discovery and development process look like?
A drug discovery program is initiated when there is a disease or clinical condition for which no suitable medical products are available or when the existing are suboptimal. Developing a new drug from the initial idea to clinical application is a complex process that follows well-established, predefined pipelines to make sure that drugs are safe and effective when they reach the public (Hughes et al., 2011).
Finding new drugs usually consists of five main stages (Singh et al., 2023)
- A pre-discovery stage in which basic research is performed to understand the mechanisms that lead to diseases, to gain new insights, and to propose possible targets against which a drug can be designed to stop or reverse the effects of the disease.
- The drug discovery stage, during which scientists search for molecules, usually small molecules and biologics or alternative therapeutic strategies that may interfere with or cure the disease under investigation or at least alleviate the symptoms.
- The preclinical development stage, which focuses on clarifying the mode of action of the drug candidates, investigating toxicity (side effects or adverse effects), validating efficacy in various in vitro and in vivo models, determining the best dosage and route of administration, and starting to evaluate the formulation.
- The clinical stage, in which the drug candidate is studied in humans, such as how it affects different groups of people (e.g., by gender, race, or ethnicity), how it interacts with other drugs and treatments, and how is its effectiveness compared with similar drugs.
- The review, approval, and post-market monitoring stage during which the drug is approved (or not).
The importance of RNA-Seq in drug discovery
The transcriptome is the complete set of RNA molecules in a cell, and while genomics provides a static view of an organism’s genome, transcriptomics reveals the dynamic landscape of gene expression. Without understanding this dynamic, one cannot understand disease mechanisms, drug responses, and inter-patient variability. At the forefront of next-generation sequencing technologies, RNA-Seq has rapidly evolved to become an invaluable tool for transcriptome profiling and gene expression analysis, and thus critical for the development of next-generation therapies. RNA-Seq helps to discover new functional genes, provides insight into the spatial and temporal expression of specific tissue or cellular genes, enables the exploration of small RNAs, all of which is widely used nowadays in disease diagnosis, drug screenings, etc. Besides bulk RNA-Seq, single-cell RNA-Seq has an immense application in drug discovery and development, especially in cancer research and development of cancer therapeutics. Our future blog articles will cover the topic of single-cell RNA-Seq and cancer drug discovery in more detail.
Now, let’s look at some applications of RNA-Seq in drug discovery and development!
RNA-Seq helps discover molecular mechanism of diseases
RNA-Seq detects differentially expressed transcripts and can reveal new molecular mechanisms of disease. Discovering the molecular mechanisms of disease is an important prerequisite for the development of new drug targets. For example, RNA-Seq has helped identify distinct oncogene-driven transcriptome profiles, thus enabling the identification of potential targets for cancer therapy (Yang et al., 2020).
RNA-Seq in target discovery and selection
Identifying potential drug target genes is a critical, but challenging step in drug development. RNA-Seq is at the heart of this process, helping to uncover genes and pathways that play an important role in disease. Once a drug has been selected for further study, RNA-Seq can also be used to detect drug-induced genome-wide changes in gene expression (Yang et al., 2020). At this point of research, understanding primary and secondary drug targets is of great importance, as it contributes greatly to the potential development of novel therapeutics. This is where one application of RNA-Seq, time-resolved RNA-Seq, comes into play – as a typical RNA-Seq approach cannot properly differentiate between primary and secondary drug effects (see: The importance of time-resolved RNA sequencing in the drug discovery process).
RNA-Seq in biomarker discovery
Biomarkers are indicators that can reveal the presence, progression, or a severity of a disease, not only aiding in early diagnosis but also serving as potential targets for drug development. RNA-Seq is a powerful tool for identifying biomarkers, because by analyzing the transcriptome, researchers can pinpoint genes whose expression profiles correlate with specific diseases. For example, in cancer research, RNA-Seq has proven to be invaluable in the discovery of several types of cancer biomarkers, including those that indicate cancer progression, recurrence and predict treatment response (Ergin et al., 2022).
A good example of cancer biomarkers are gene fusions, which drive malignancy, and are therefore promising new targets for personalized therapies. Fusion genes have been reported as therapeutic targets in acute myeloid leukemia; recurrent gene fusions, key drivers of cancer, have been discovered in breast and colorectal cancer. Expression of gene fusions can be detected by whole transcriptome sequencing, although in clinical samples they are primarily detected by the RNA-CaptureSeq approach, a targeted RNA sequencing method that can achieve greater sequencing depth for selected transcripts. RNA-Seq has uncovered another group of cancer biomarkers, namely small RNAs, such as miRNAs, and various types of non-coding RNAs, lnRNAs, and circRNAs, all of which are present in different types of cancer (Mercer et al., 2011; Ergin et al., 2022).
RNA-Seq helps identify genes involved in drug resistance and sensitivity
Chemotherapy is currently the gold standard therapy for many types of cancer, but unfortunately, chemotherapy drug resistance is becoming a growing concern for patients as it leads to cancer treatment failure (Khatoon et al., 2014; Yang et al., 2020).
Because RNA-Seq can help determine how a disease develops, why it is affected by drugs, and identify novel transcripts and splicing events, it can also be used to identify genes associated with drug resistance. For example, triple-negative breast cancer (TNBC) is a type of breast cancer that is extremely difficult to treat. Due to the absence of three types of receptors (estrogen receptor (ER), progesterone receptor (PR), and human epidermal growth factor receptor 2 (HER2)), a high degree of molecular variability, and strong drug resistance, consistent treatment results cannot be achieved. In one study (Shaheen et al., 2018; Yang et al., 2020), RNA-Seq was used to determine and analyze the differentially expressed genes and their biological functions of two different TNBC drug-resistant cell lines after JQ1 and dexamethasone therapy, and the results showed significant differences in the expression of cytokine-cytokine receptor interaction pathways, providing new ideas for the development of drugs to treat TNBC.
The role of microRNAs (miRNAs) in the regulation of drug resistance is widely accepted. Small RNA-Seq or microRNA-Seq is a perfect tool to identify potential miRNA that drive drug resistance, as it allows the comparison of miRNA expression profiles between drug-resistant and non-resistant cells. Resistance to doxorubicin (DOX) is a common obstacle to effective treatment of liver cancer. Zhang et al. investigated the role of miRNAs in DOX resistance of hepatocellular carcinoma (HCC), a deadly virus-induced cancer with growing resistance towards DOX. Using RNA-Seq, they were able to determine which miRNAs were down-regulated in their model cell types, as well as to which functional pathway they were associated with. This study provided a general description of the miRNA expression profile, enabling to search for possible miRNAs to assist therapy to overcome DOX resistance, or to develop new drugs to avoid drug resistance, in further studies (Zhang et al., 2013).
RNA-Seq helps in the assessment of drug toxicity
Drug side effects are a major obstacle and a reason why many drugs fail in clinical trials. Various omics technologies, including transcriptomics, are being used more routinely to understand the biological mechanisms involved in drug effects. RNA-Seq helps to understand and assess drug toxicity by monitoring changes in gene expression caused by a drug, providing invaluable information on potential drug side effects, and helping to optimize drug formulations to minimize harm (Nguyen et al., 2022).
RNA-Seq in Drug Repurposing
Developing a new drug is a time-consuming and costly endeavour, so in clinical research, much efforts have been made to repurpose drugs that may have an untapped potential for treating different conditions. RNA-Seq can help these efforts by enabling transcriptome profiling and screening for therapeutic targets. A good example is a study where authors screened the therapeutic targets in adult acute myeloid leukemia (AML), a very heterogeneous condition with more than 20 subtypes and the 5-year overall survival rate of only 27 %. They interrogated 200 primary specimens with mubritinib, originally developed to treat breast cancer and could show that chemotherapy-sensitive AMLs that display transcriptomic hallmarks of hypoxia have resistance to mubritinib. They also revealed that mubritinib can cause the death of certain cancer cells (Saini et al., 2017; Yang et al., 2020).
Pharmacogenomics
Pharmacogenomics, the study of how an individual’s genes affect their response to drugs, relies heavily on RNA-Seq. By examining gene expression patterns, researchers can optimize drug dosages to increase treatment efficacy while minimizing side effects (Khatoon et al., 2014; Xu et al., 2016; Yang et al., 2020).
The Importance of time-resolved RNA sequencing in the drug discovery process
Conventional RNA-Seq can only capture a snapshot of the transcriptome at the time of sampling, without the added dimension of time. And while conventional RNA sequencing allows for comprehensive steady-state transcriptome analyses, its ability to probe for transcriptional responses to various cell perturbations, such as drug treatments, is limited by the diversity of messenger RNA (mRNA) and protein half-lives (Herzog et al., 2017).
Suppose we are designing an experiment to test the effect of a drug on the transcriptome of cells, and we need to choose a time point to sample after the drug treatment. If we choose an early time point, detectable changes in expressed transcripts are inevitably biased toward short-lived ones, while sampling at later time points does not enable us to distinguish direct from indirect effects, or in another words, primary from secondary drug effects. This is where time-resolved RNA-Seq comes into play.
Time-resolved RNA-Seq or time-resolved RNA profiling is an application of RNA-Seq that allows for observations of RNA abundances over time in biological samples, enabling investigation of RNA kinetics.
Incorporation of information on the timing of expression of different transcripts can be a real game-changer for drug discovery and development pipelines as it allows the distinction between primary (direct) and secondary (indirect) effects of the tested drug(s). This helps to resolve complex regulatory networks and predictions of combinatorial effects to increase efficacy.
High-throughput kinetic RNA sequencing with SLAMseq
SLAMseq method (Thiol (SH)-Linked Alkylation for the Metabolic sequencing of RNA) is a method developed by the Ameres lab (Herzog et al., 2017) that allows the identification and quantification of newly synthesized (nascent) and existing RNA from the same sample in parallel, without the need for biochemical isolation.
In combination with RNA-Seq, SLAMseq enables high-throughput kinetic RNA sequencing, or high-throughput time-resolved RNA sequencing, ideal for studying the effects of fast-acting drugs or drug candidates on RNA kinetics in cell culture experiments. Kinetic RNA-Seq assesses anabolic (RNA synthesis) or catabolic (RNA degradation) processes in a time-resolved manner, revealing significant cause-and-effect relationships of therapeutic compounds at much higher resolution than conventional methods.
Why is SLAMseq ideal for drug discovery and testing?
SLAMseq measures RNA synthesis and RNA degradation separately allowing the capture of nascent RNA expression or transcript stability in response to drug administration. With the added dimension of time, primary or secondary transcriptional targets are identified, and off-targets detected. Understanding the full range of drug effects helps to predict potential side effects, investigate drug-drug interactions and optimize efficacy. SLAMseq has been used extensively for cancer research (Paris et al., 2019; Zhang et al., 2022) and is applicable to fast-acting drugs, including small molecules, rapid RNA therapeutics, and biologicals.
The ability to differentiate between primary and secondary targets with SLAMseq is beneficial at multiple stages of drug development, including target identification, potential target validation, and lead optimization. SLAMseq provides an unbiased picture of transcriptional processes in the living cell and adds value to any drug discovery journey.
High-throughput kinetic RNA sequencing with SLAMseq reveals drug mechanisms of action and thus supports the identification of potential biomarkers, that can indicate the presence or severity of disease, and are becoming the driving force behind personalized treatments and monitoring of disease progression. By supporting biomarker identification, SLAMseq can accelerate the development of new therapies and improve patient outcomes.
How does SLAMseq work?
The SLAMseq method is based on the metabolic labeling of RNAs with 4-thiouridine (S4U), a nucleotide analog that is readily imported into cells and incorporated into RNA, combined with thiol(SH)-linked alkylation using thiol-reactive compound iodoacetamide (IAA), which modifies the 4-thiol group of S4U-containing nucleotides via the addition of a carboxyamidomethyl group. Lexogen’s SLAMseq combines the patented S4U labeling technology with a streamlined workflow in the only commercially available kit to enhance and accelerate your large-scale drug discovery.
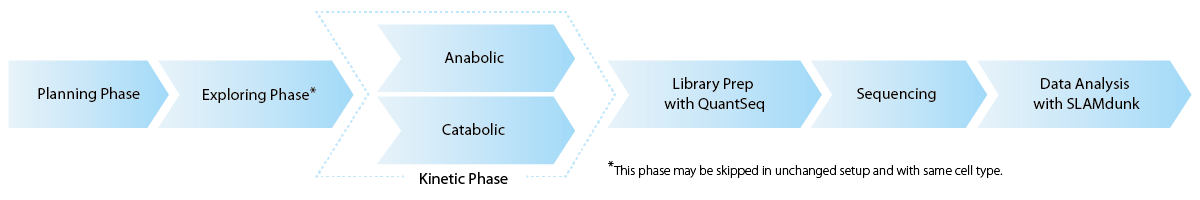
RNA labeling is specific to each cell type and culturing method. For every new experimental setup, SLAMseq requires optimization of labeling conditions and efficiency testing of S4U uptake (Exploring phase). For RNA kinetics analysis, either RNA synthesis (anabolic) or RNA degradation (catabolic) rates are measured and monitored to generate treatment response profiles. Each sample is prepped with QuantSeq 3’ mRNA-Seq, sequenced and data analyzed using the SLAMdunk analysis pipeline, a modified alignment algorithm specially adapted to report read counts for T>C-containing reads for downstream analysis (Herzog et al., 2017) (Fig. 1).
Figure 1 | Most important steps for High-Throughput Kinetic RNA Sequencing with SLAMseq.
SLAMseq laboratory experimental workflow
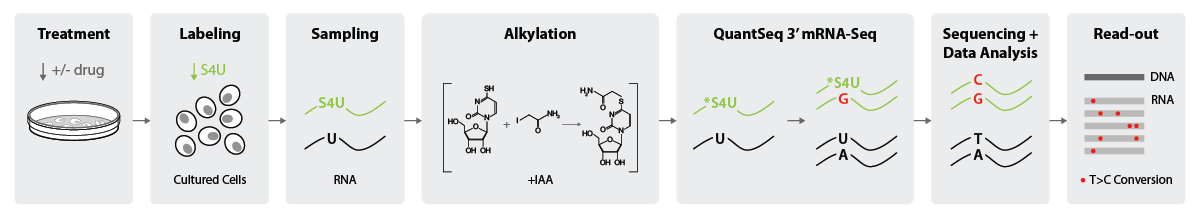
SLAMseq is designed to be used with cultured cells (Herzog et al., 2017; Muhar et al., 2018). To assess RNA synthesis, cultured cells are first treated with a drug or a control, followed by the addition of 4-thiouridine (S4U) to the culturing media. S4U will be incorporated into newly synthetized transcripts, thereby labelling them. At specific, previously selected time points, cells are collected, RNA is isolated (Sampling step) and alkylated with Iodoacetamide (IAA). RNA-Seq libraries are generated with QuantSeq 3’ mRNA-Seq resulting in a nucleotide conversion during reverse transcription, as reverse transcriptase will incorporate a G instead of an A at positions where reduced *S4U-modified nucleotides are encountered. Sequencing will detect thymine-to-cytosine (T>C) mutations in S4U-labeled transcripts, and these will enable bioinformatic analysis of changes in nascent RNA levels (Fig. 2). For a visual representation of this workflow, check out our RNAEXPERTise animated video on SLAMseq.
Figure 2 | Simplified lab workflow for assessing RNA synthesis High-Throughput kinetic RNA sequencing with SLAMseq combined with QuantSeq 3′ mRNA-Seq.
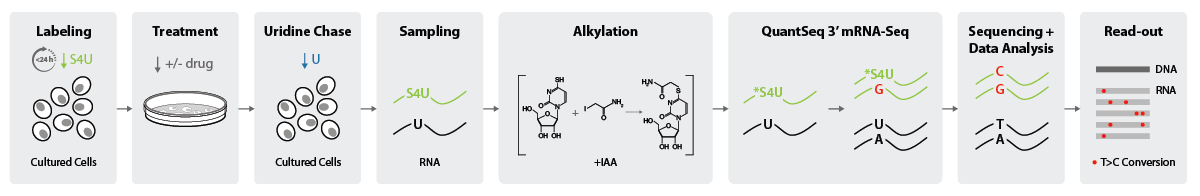
For assessing RNA turnover, the upstream part of the experiment (until Sampling) differs. Here, a pulse-chase approach is used, and cells are first labeled with S4U for up to 24 h until saturation is reached. As a next step, the treatment will be applied and after incubation time, Uridine will be introduced as a chase. The S4U signal will, consequently, decrease over time (Figure 3).
Figure 3 | Simplified lab workflow for High-Throughput kinetic RNA sequencing for assessing RNA turnover with SLAMseq combined with QuantSeq 3′ mRNA-Seq.
Summary
RNA-Seq has revolutionized our understanding of the highly dynamic nature of the transcriptome and has become a virtually irreplaceable tool in biological, medical, or clinical studies. From providing insight into differential gene expression in response to treatments, to investigating alternative splicing, isoforms, gene fusions, small RNAs, post-transcriptional modifications, single-cell transcriptomics with scRNA-Seq, to elucidating RNA kinetics, RNA-Seq and its many sub-technologies have the potential to identify novel disease biology, profile biomarkers, make diagnoses – and as we outline in this blog article – significantly complement drug discovery and development. RNA-Seq is used at various stages of drug discovery and development and is used by researchers to:
- Understand the molecular mechanism of diseases.
- Identify potential drug target genes.
- Discover biomarkers.
- Identify genes involved in drug resistance and sensitivity.
- Assess drug toxicity.
- Repurpose drugs.
- Understand how an individual’s genes affect their response to drugs (pharmacogenomics).
- Develop personalized therapies.
To dive even deeper, and really understand the intrinsic details of the direct and indirect effects of drugs, one can rely on time-resolved RNA sequencing, using the well-established SLAMseq technology for high-throughput kinetic RNA sequencing.
Interested in learning more about SLAMseq and time-resolved RNA profiling? Check out SLAMseq product page and Lexogen’s webinar with Dr. Stephan Ameres on Time-resolved RNA profiling for cancer research.
References:
Written by Masa Ivin, PhD