QuantSeq 3’ mRNA-Seq Library Prep Kit FWD
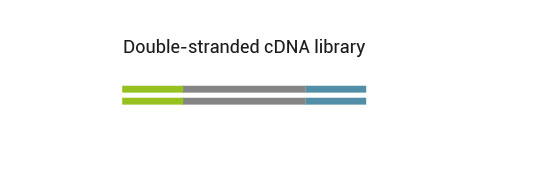
The QuantSeq FWD Kit is a library preparation protocol designed to generate Illumina compatible libraries of sequences close to the 3’ end of polyadenylated RNA.
Dear Customer, QuantSeq V1 kits will be phased out soon. The last order date is October 31, 2023.
For more information, please get in touch with us at info@lexogen.com or sales@lexogen.com. Thank you!
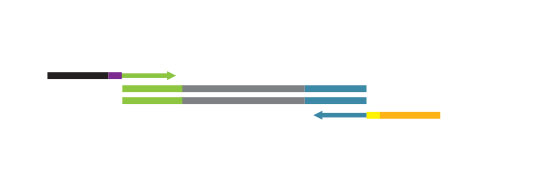
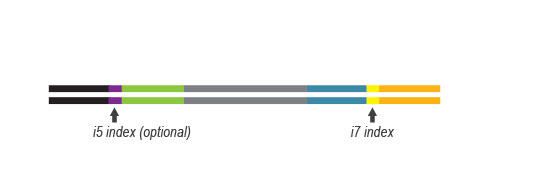
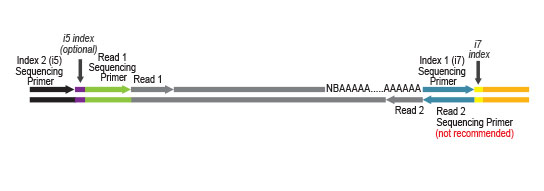
QuantSeq FWD contains the Illumina Read 1 linker sequence in the second strand synthesis primer, hence NGS reads are generated towards the poly(A) tail, directly reflecting the mRNA sequence (see workflow). This version is the recommended standard for gene expression analysis.
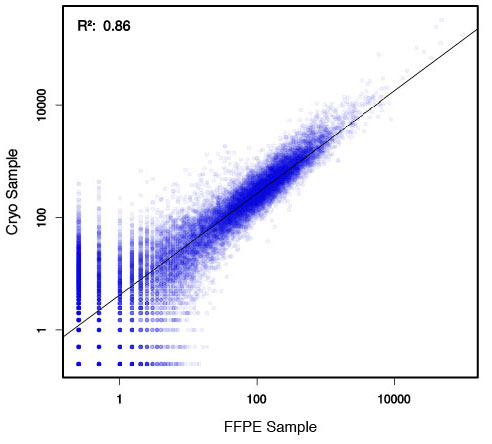
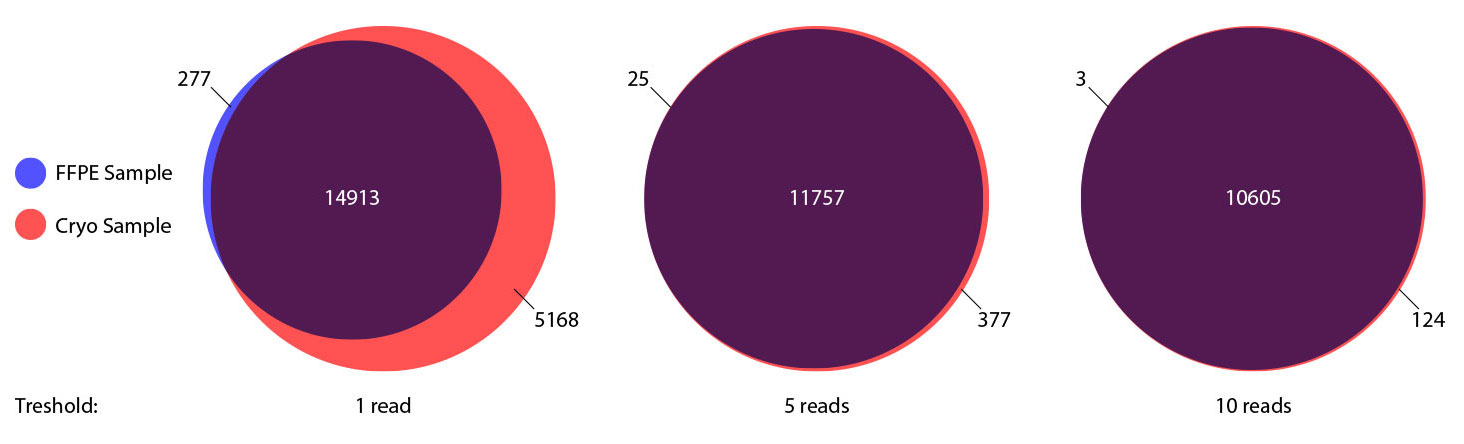
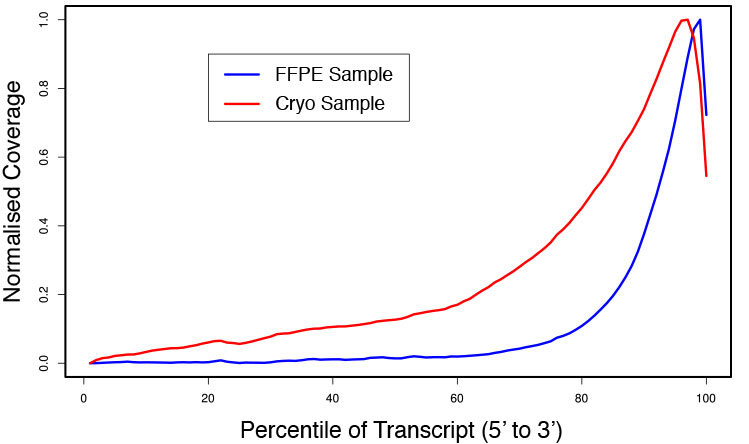
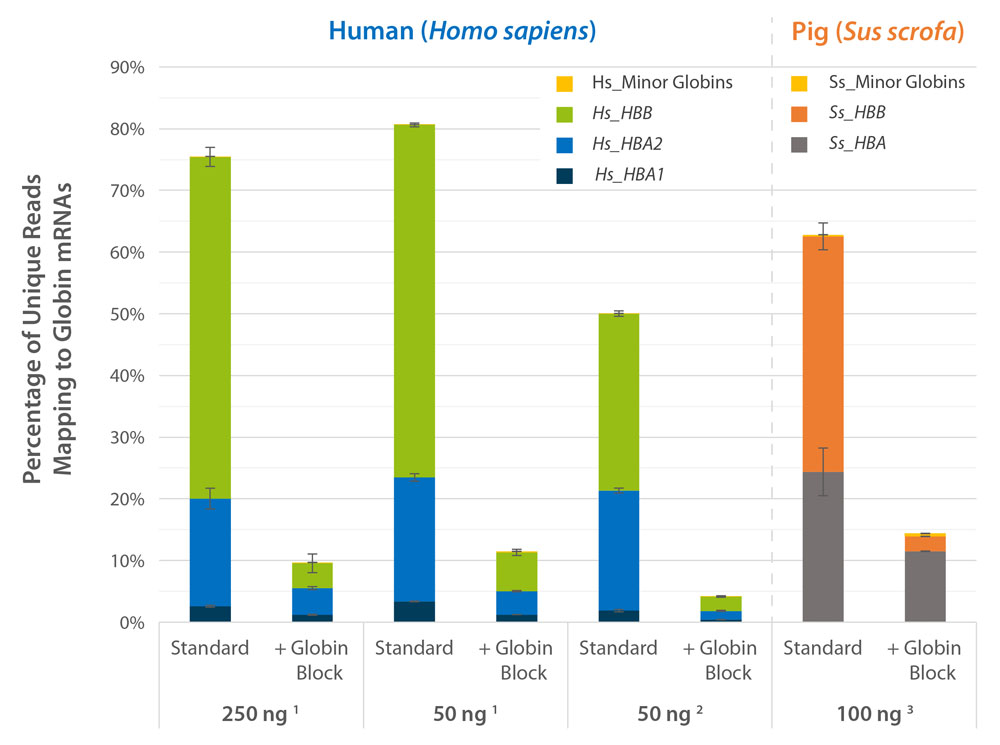
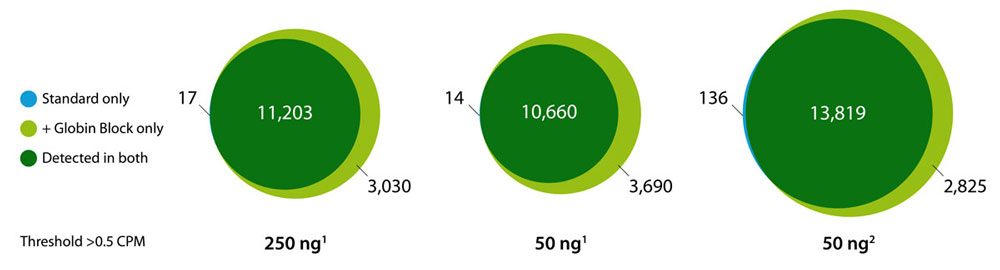
Performance
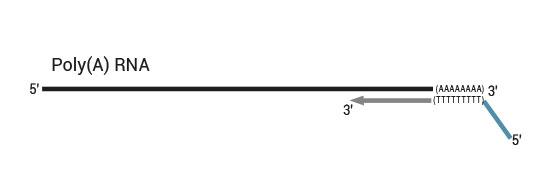
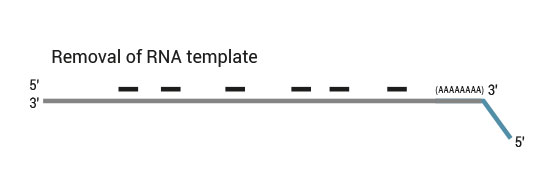
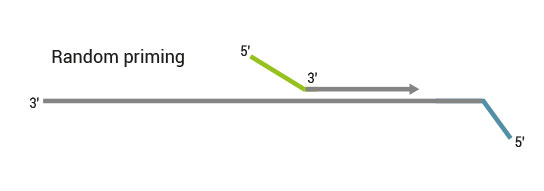
Workflow
QuantSeq has a short and simple workflow and can be completed within 4.5 hours. The required hands-on time is less than 2 hours. The kit uses total RNA as input, hence no prior poly(A) enrichment or rRNA depletion is needed.
Featured Publications
Automation
Automated QuantSeq protocol for 3’ mRNA-Seq Library Preparation
Automating the process of library preparation has the advantage of avoiding sample tracking errors, dramatically increasing throughput, and saving hands-on time.
QuantSeq library preparation and has been successfully implemented on several liquid handlers:
- Perkin Elmer: Sciclone® / Zephyr®
- Hamilton: Microlab STAR / STARlet
- Agilent: NGS Workstation (NGS Bravo Option B)
- Beckman Coulter: Biomek FXP, Biomek i5, Biomek i7
- Eppendorf: EpMotion® 5075
- Opentrons® OT-2
QuantSeq automation on other platforms may also be possible. Please contact support team for more information.
Two key parameters must always be considered when automating a protocol:
- volume optimization
- script compatibility
In some instances, our kits will provide enough reagents while, in other instances, you will need a higher reagent volume or script adjustments. At Lexogen, we will help you find the best solution, tailored to your needs.
Before starting any new project involving automation, please reach out to our experts at support@lexogen.com
Please, also consider liaising with the robotic platform support team – they will be able to share the most recent version of the script.
Lexogen gladly supports the implementation of Lexogen-manufactured kits on liquid handlers, but not hardware or software issues linked to the original liquid handling instrument supplier.
Related resources:
![]() Application Note for Automation of QuantSeq 3’ mRNA Kit on the Agilent NGS Workstation
Application Note for Automation of QuantSeq 3’ mRNA Kit on the Agilent NGS Workstation
![]() Application Flyer for Automation of QuantSeq 3’ mRNA Kit on Beckman Biomek NGS Workstation
Application Flyer for Automation of QuantSeq 3’ mRNA Kit on Beckman Biomek NGS Workstation
![]() Webinar: Automation of RNA-seq: from model organisms to the clinic
Webinar: Automation of RNA-seq: from model organisms to the clinic
FAQ
Frequently Asked Questions
Access our frequently asked question (FAQ) resources via the buttons below.
Please also check our General Guidelines and FAQ resources!
How do you like the new online FAQ resource? Please share your feedback with us!
Downloads
![]() QuantSeq Product Flyer – update 04.01.2023
QuantSeq Product Flyer – update 04.01.2023
![]() QuantSeq Application Note (Nature Methods, December 2014) – external link
QuantSeq Application Note (Nature Methods, December 2014) – external link
![]() Application Note for QuantSeq – update 04.01.2023
Application Note for QuantSeq – update 04.01.2023
![]() Application Note for QuantSeq Data Analysis with PARTEK® FLOW®
Application Note for QuantSeq Data Analysis with PARTEK® FLOW®
![]() Application Note for Globin Block Modules
Application Note for Globin Block Modules
QuantSeq 3′ mRNA-Seq Library Prep Kit FWD for Illumina
![]() User Guide – update 24.01.2023
User Guide – update 24.01.2023
![]() User Guide HT – update 25.01.2023
User Guide HT – update 25.01.2023
![]() User Guide for BC1 Block Module for QuantSeq – update 25.01.2023
User Guide for BC1 Block Module for QuantSeq – update 25.01.2023
For QuantSeq Add-on Module User Guides click here
![]() User Guide for QuantSeq 3’ mRNA-Seq V2 Library Prep Kit FWD with UDI 12 nt Set (version 2) – update 12.07.2023
User Guide for QuantSeq 3’ mRNA-Seq V2 Library Prep Kit FWD with UDI 12 nt Set (version 2) – update 12.07.2023
![]() User Guide for QuantSeq 3’ mRNA-Seq Library Prep Kit FWD with UDI 12 nt Set (version 1) – update 25.01.2023
User Guide for QuantSeq 3’ mRNA-Seq Library Prep Kit FWD with UDI 12 nt Set (version 1) – update 25.01.2023
![]() User Guide for Lexogen 12 nt Unique Dual Indexing Add-on Kit V2 (version 2) – release 12.10.2022
User Guide for Lexogen 12 nt Unique Dual Indexing Add-on Kit V2 (version 2) – release 12.10.2022
![]() User Guide for Lexogen 12 nt Unique Dual Indexing Add-on Kit (version 1) – update 25.01.2023
User Guide for Lexogen 12 nt Unique Dual Indexing Add-on Kit (version 1) – update 25.01.2023
![]() PCR Add-on Kit for Illumina User Guide – update 03.01.2023
PCR Add-on Kit for Illumina User Guide – update 03.01.2023
![]() Lexogen i5 6 nt Dual Indexing Add-on Kits (5001-5096) User Guide – update 03.01.2023
Lexogen i5 6 nt Dual Indexing Add-on Kits (5001-5096) User Guide – update 03.01.2023
![]() Lexogen UDI 12 nt Unique Dual Index Sequences – update 26.03.2021
Lexogen UDI 12 nt Unique Dual Index Sequences – update 26.03.2021
![]() Lexogen i7 and i5 Index Sequences – update 05.05.2020
Lexogen i7 and i5 Index Sequences – update 05.05.2020
autoQuantSeq 3′ mRNA-Seq Library Prep Kit for Illumina
- Perkin Elmer: Sciclone® / Zephyr®
- Hamilton: Microlab STAR / STARlet
- Agilent: NGS Workstation (NGS Bravo Option B)
- Beckman Coulter: Biomek FXP, and Biomek i7
- Eppendorf: EpMotion® 5075
Please inquire at info@lexogen.com for the automation scripts
Safety Data Sheet
If you need more information about our products, please contact us through support@lexogen.com or directly under +43 1 345 1212-41.
QuantSeq Bioinformatics Data Analysis
Find more about the QuantSeq Data Analysis here.
Use this command line tool for demultiplexing QuantSeq-Pool data – Demultiplexing and Error Correction Tool – iDemux
Buy from our Webstore
Need a web quote?
You can generate a web quote by Register or Login to your account. In the account settings please fill in your billing and shipping address. Add products to your cart, view cart and click the “Generate Quote” button. A quote in PDF format will be generated and ready to download. You can use this PDF document to place an order by sending it directly to sales@lexogen.com.
Web quoting is not available for countries served by our distributors. Please contact your local distributor for a quote.