Lexogen now offers CORALL mRNA-Seq bundles including poly(A) selection providing a convenient complete workflow solution for mRNA-Seq library preparation.
Features and Benefits
CORALL mRNA-Seq features an easy all-in-one workflow from total RNA to ready-to-sequence libraries and takes less than 6 hours to complete. The kit is applicable for a wide range of total RNA input (from 1 ng to 1 µg) and comes with Unique Molecular Identifiers (UMIs) and Unique Dual Indexing (UDIs).
UMIs are introduced during CORALL library generation and enable the removal of PCR duplicates for precise quantification.
CORALL mRNA-Seq Kits are available with up to 384 pre-mixed UDIs for Illumina’s Forward Strand (A) and Reverse Complement (B) workflows (Set A1-A4: Cat. No. 158-161, 163, or Set B1: Cat. No. 162). Lexogen’s new 12 nt UDIs feature superior error correction for maximal sequencing data output especially at high-level multiplexing, for example, on NovaSeq® instruments.
Reads derived from CORALL libraries are highly strand-specific and correspond directly to the mRNA sequence. Furthermore, Read 1 contains the UMI information making it directly accessible in cost-efficient single-read sequencing mode. User-friendly data analysis pipelines are available on various platforms for CORALL users to explore their data – even without proficiency in bioinformatics.
The complete CORALL mRNA-Seq workflow is automation compatible and can hence be easily implemented in projects where high throughput sample processing is required.
CORALL mRNA-Seq as well as CORALL Total RNA-Seq (in combination with ribosomal RNA depletion) offer excellent gene detection and transcript coverage even for low RNA input amounts (between 1-10 ng) which are in line or outperform other leading protocols on the market.
Transcript / Gene Detection for various input amounts at different read depth
CORALL mRNA-Seq consistently detects between 68,000 and 70,000 transcripts across a wide range of total RNA input (1 ng – 100 ng prior to poly(A) selection, Fig. 1) corresponding to ~20,000 genes.
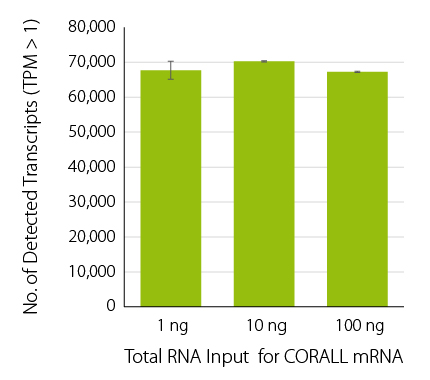
Figure 1 | Consistent transcript detection across a wide range of input amounts. No. of detected transcripts for CORALL mRNA-Seq from 1 ng, 10 ng, and 100 ng total RNA input at a read depth of 10 M reads / sample and a threshold of >1 TPM (Transcript Per Million).
Coverage uniformity across input amounts
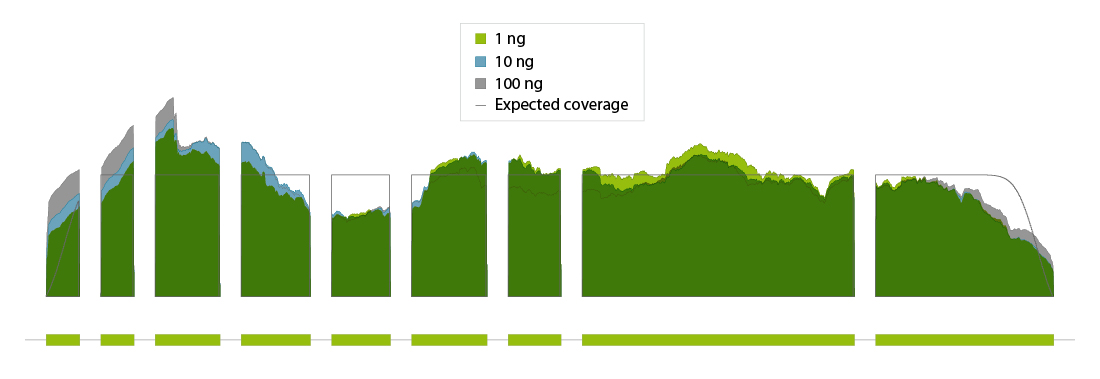
Coverage profiles show uniformity across the transcript body regardless of the input RNA amount used for poly(A) selection emphasizing consistent performance and suitability of CORALL mRNA-Seq even for low input applications (Fig. 2).
Figure 2 | Uniform coverage of the GAPDH transcript over a broad range of RNA input amounts. CORALL mRNA-Seq using 1 ng, 10 ng, and 100 ng RNA input prior to poly(A) selection. Mapped reads are visualized in condensed coverage profiles of the entire transcript. Generic introns are shortened in the visualization to gaps of equal length.
Reproducibility across replicates and consistent transcript expression across input amounts
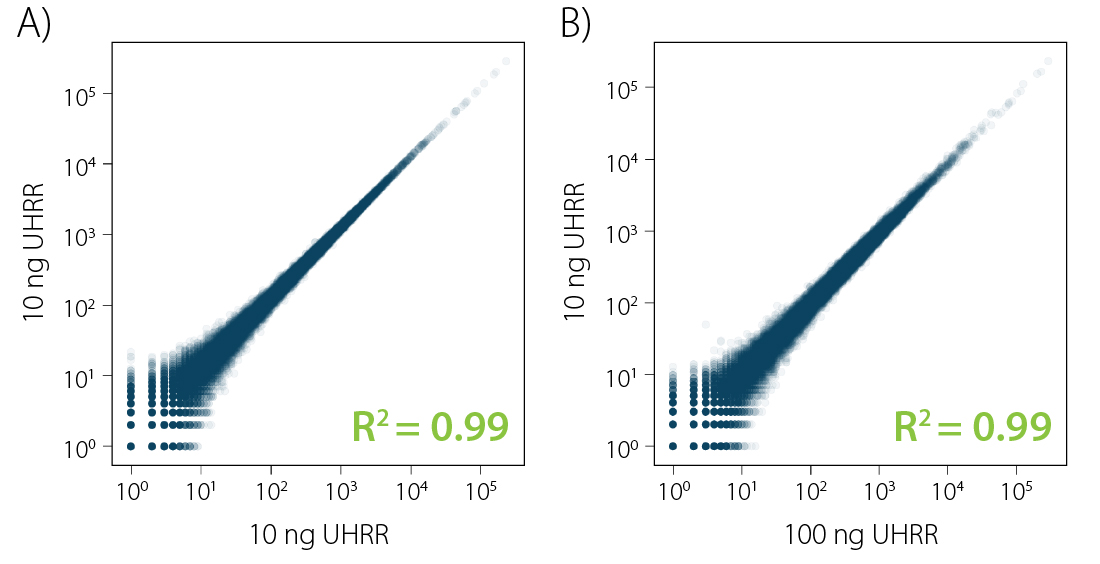
Correlation analysis comparing individual replicates for CORALL mRNA-Seq reveals excellent reproducibility (Fig. 3 A). Additionally, CORALL mRNA-Seq maintains consistent transcript expression across input amounts (Fig. 3 B).
Figure 3 | Excellent reproducibility and consistent transcript abundance across input amounts. Correlations between A) replicates for CORALL mRNA-Seq from 10 ng RNA and B) transcript abundance from CORALL mRNA samples using 100 ng vs. 10 ng RNA as input for poly(A) selection.
Learn more about CORALL mRNA-Seq Library Prep
Lean more about Lexogen 12 nt Unique Dual Index System