Formalin-fixed paraffin-embedded (FFPE) human tissue samples provide information for the study of disease development, progression, and treatment efficacy.
One of the largest research applications for FFPE samples is in oncology and cancer research1. Patient biopsies in the form of FFPE samples are necessary to stage and characterize cancers, establish a cancer diagnosis, select treatment options, and estimate treatment efficacy.
Cancer research involves identifying and understanding the cellular and molecular components of tumor cells. It also involves discovering the reasons why cells become tumor cells. To make these discoveries, researchers need access to highly informative samples, including FFPE samples.
FFPE samples are extensively used in immunology2,3 to study immune cells in various healthy and diseased states. Good example is again cancer, as cancer immunology, or immuno-oncology, a branch of immunology that is concerned with understanding the role of the immune system in the progression and development of cancer, benefits a lot from investigating interplay between immune cells and cancer cells in FFPE samples. By studying FFPE samples from a person with an autoimmune disease4, it is possible to determine the cause and develop drugs against the disease. FFPE samples are also used for infectious diseases studies, for example, for the detection of different pathogens in the tissues.
FFPE tissues are important tools in hematology5 to study blood-related disorders. Studying bone marrow with FFPE samples can be applied to a variety of different fields of study, such as genetics, tissue regeneration, and toxicology. Brain FFPE samples are crucial for studying neurodegenerative disorders such as Alzheimer’s6.
FFPE samples are not only important for understanding individual cases, but they also contribute to collective knowledge about diseases. Knowledge obtained from studying these samples can lead to better treatments, improved diagnostics, and even preventive measures for future generations.
Methods to study FFPE samples
Proteins, DNA, and RNA are preserved in the tissue thanks to formalin, a fixative for preparing FFPE samples. Several methods are suitable for studying proteins and nucleic acids captured in FFPE samples.
NOTE: Before conducting any downstream study, it is necessary to obtain sections from FFPE tissue blocks. The standard thickness for FFPE tissue sections is 3-5 micrometres (or microns), which is approximately the thickness of a cell. Slides with 3–5-micron sections are commonly used for staining, including the standard hematoxylin & eosin (H&E) stain, special stain, or stain for immunohistochemistry (IHC). Additionally, there are thicker sections known as FFPE curls, which are 10 microns or greater in thickness. FFPE curls are generally used when extracting RNA or DNA from the FFPE samples.
Histo-pathology on FFPE samples
Standard histopathological analysis of tissues begins from FFPE blocks that are sectioned and mounted on glass slides. They can be stained with a standard H&E stain to observe tissue structure, infiltration of immune cells, inflammation, and potentially transformed cells, to name a few (Figure 1). Immunohistochemistry (IHC) is used to identify specific proteins. In IHC, tissue sections are bathed in a solution containing antibodies that will bind to specific proteins or structures. Stains are then used to visualize the antibodies, showing the samples’ specific protein and location.
IHC on FFPE samples is widely used in oncology to diagnose, evaluate prognosis, and predict response to therapy. FFPE samples for IHC allow clinicians to monitor immune cell infiltration, essential for treatment efficacy in cancer.

Next-generation sequencing on FFPE samples
Clinical biopsy tissue samples come in many forms; some are more suitable for next-generation sequencing (NGS) studies than others. Fixing tissues in formalin and embedding in paraffin (FFPE) and snap-freezing fresh tissue are the most common methods for human tissue preservation7. The FFPE approach uses formalin to stop and preserve cellular processes, whereas fresh frozen sample preparation relies on rapid cooling-down to achieve the same goal. Both approaches have pros and cons; for example, frozen tissue collections cannot be stored for as long as FFPE tissues, and maintenance is way more costly and demanding, while nucleic acids isolated from frozen samples are of better quality. Choosing one or another will ultimately depend on the study’s requirements and technical conditions. While in this article we focus on FFPE samples in NGS studies, we will dive deeper into the comparison between fresh frozen and FFPE samples in NGS studies in our follow-up blog article.
The fixation in formalin preserves nucleic acids, so DNA and RNA can be extracted from FFPE tissues. FFPE samples, therefore, serve as an excellent material for genomic, epigenetic, and transcriptomic studies, using next-generation whole genome sequencing (DNA-Seq), bulk RNA-Seq, and even single-cell RNA-Seq or single-nucleus RNA sequencing8,9(Figure 2).

For example, in oncology and cancer research, the analysis of DNA and RNA from FFPE samples allows clinicians to understand why a certain treatment worked for one patient and not another, why the patient developed resistance to the treatment and what mutations, or transcriptome changes are associated with that resistance. Even though NGS is widely implemented, there are not many official recommendations for the use of NGS in oncology coming from scientific societies. One of the first came from The European Society for Medical Oncology (ESMO), where NGS is recommended for use on tumor samples in advanced non-squamous non-small-cell lung cancer (NSCLC), prostate cancers, ovarian cancers, and cholangiocarcinoma10.
NGS has been used in many cancer research and clinical studies. In addition, genomic and transcriptional studies from FFPE samples have the potential to contribute significantly to personalized medicine, as frozen fresh samples already do. For example, frozen tissues were used to determine a 70 gene signature in breast cancer, which allowed clinicians to determine if patients could undergo conventional chemotherapy treatments11. Based on the results of the clinical trial, researchers concluded that nearly half of women who were categorized as having “high clinical risk” may not have required chemotherapy. This insight increased the personalization of the treatment regimen, meaning that individuals did not need harmful treatments with many side effects.
Globally, between 50 and 80 million FFPE samples are potentially suitable for NGS analysis, and that is for solid tumors alone12. So far, the biggest bottleneck to performing NGS from FFPE samples is obtaining high-quality DNA and RNA owing to chemical modifications, fragmentation of DNA and RNA, and cross-linking to proteins introduced through the fixation process. Developing and validating optimized protocols for FFPE tissue and implementing state-of-the-art DNA and RNA sequencing technologies are necessary to unlock the precious information within these biobank samples and fuel clinical research and the fight against cancer, neurodegenerative and other human diseases.
DNA-Seq on FFPE samples
Many key studies have used DNA-Seq to analyze FFPE-DNA, and sequencing is also officially recommended to diagnose some cancer types10. We will outline some of these studies to demonstrate the value of FFPE samples for sequencing studies and explain which different types of DNA-Seq can be performed on FFPE-DNA13.
DNA-Seq can be used to determine the sequence of specific genes, or parts of them (targeted sequencing via targeted panels), to sequence all the coding regions in the genome (whole-exome sequencing, WES), or to sequence the entire genome, including intronic and intergenic regions (whole-genome sequencing, WGS)14.
Targeted (direct) sequencing focuses on a selected set of genes or genomic regions. In cancer research, this method is used when the association of specific genes with specific cancer is already known. To facilitate targeted cancer FFPE sequencing studies, many providers offer custom panels where researchers can select their amplicons of interest. Researchers have successfully used FFPE-DNA for targeted sequencing to analyze copy number variants and detect mutations15,16. Among the examples are a study where targeted DNA-Seq assessed KRAS mutations (a known mutation causing resistance to therapy) in FFPE samples of metastatic colorectal cancer or a study where more than 180 cancer-associated genes were analyzed in FFPE samples of patients with advanced prostate cancer15.
Whole genome sequencing is a method for analyzing entire genomes, and is used as a “hypothesis-free approach” where novel genome events can be identified, without a priori knowledge of effected targets. With the progress in technologies and cost reduction, WGS has more and more advantages over targeted sequencing, especially in clinical cancer research studies17. WGS is especially valuable if all variant types, including single-nucleotide variants (SNVs), small insertions/deletions (indels), and structural variants such as copy-number alterations (CNAs) are of research interest18. Fresh frozen tissue is the optimal source of DNA for WGS of cancer patients. However, fresh frozen tissue is only occasionally available, limiting the widespread application of WGS in clinical practice. This is where FFPE samples come into the picture. A prospective study, a pilot study for the 100,000 Genomes Project, explored the viability of using FFPE samples as a source of DNA for clinical WGS in cancer patients. Even though shortcomings of FFPE samples compared to fresh frozen were identified, they could be minimized by careful DNA extraction and bioinformatics analysis, supporting the use of FFPE cancer samples in clinical settings18.
FFPE samples are useful also for whole exome sequencing, as shown in the study where WES was performed on samples of gastrointestinal stromal tumors (GIST). The authors could easily select FFPE samples of high quality that yield sequencing results comparable to the fresh frozen counterparts19.
Challenges when performing DNA-Seq from FFPE-DNA
Even though there are a lot of protocol optimizations specifically developed for DNA-Seq from FFPE samples, it is important to be aware of some of the challenges when working with FFPE-DNA. Low input amounts, non-uniform ends, crosslinking to proteins and formalin-caused DNA modifications are common and can hinder library preparation and downstream analysis20.
While formalin is very effective at storing and preserving DNA within tissue samples, it does create various types of DNA modifications and damage. Modifications in FFPE-DNA mainly occur randomly but more frequently in AT-rich genomic regions, leading to a higher prevalence of GC-rich sequences than in DNA isolated from fresh tissues12.
A common form of DNA damage caused by formalin fixation is DNA fragmentation, where the backbone of the DNA macromolecule is cleaved into separate segments. DNA fragmentation often happens after prolonged storage and varying environmental conditions during the storage21. It can result in low amounts of suitable templates for PCR amplification22.
Cytosine deamination is one of the most common chemical alterations found in FFPE-DNA, occurring due to spontaneous deamination of cytosine. In living cells, enzymes glycosylases are responsible for repairing these events, but in the FFPE tissues, glycosylases are inactivated by the fixation23. Deaminated cytosine results in uracil, which instead of guanine pairs with adenine. When cytosine gets methylated (5-methylcytosine), its deamination leads to thymine which also pairs with adenine. Both cases lead to the base pair alteration C>T/ G>A in the newly synthesized strand, resulting in a single base substitution artefact and false signals12.
Further DNA alterations caused by fixation with formalin include base modification24 resulting in a base with altered base-pairing abilities. These modified bases can often further react to form a covalent cross-link with another nucleophilic group in their proximity. These cross-links can change base-pairing characteristics, leading to the incorporation of non-complementary nucleotides in daughter strands during sequencing library preparation. In addition, cross-links can also induce polymerase “blocks” during PCR25.
In addition, fixation with formalin can lead to the generation of apurinic/apyrimidinic (AP) sites within the double strand (so-called base excision) by hydrolysis of the N-glycosylic bonds21. AP sites are more prone to damage and fragmentation. Regarding DNA-Seq of such DNA molecules, they may not be amplified sufficiently for sequencing. This results in a reduced diversity of functional sequencing library molecules, reducing the library complexity and causing loss of information.
Tips to minimize or mitigate the impact of FFPE-DNA alterations on the success of DNA-Seq
In a review article titled “A critical spotlight on the paradigms of FFPE-DNA sequencing”, the authors have covered many important aspects of performing DNA-Seq on FFPE-DNA, challenges, workarounds, and considerations to be mindful of, all up to bioinformatics tools. The value of using FFPE samples for genome sequencing is so high, especially in cancer research, that investing in these workarounds really pays off, as it enables the best possible quality of data.
- Taking care when performing DNA extraction
FFPE-DNA extraction is critical and should be performed with caution, ideally using protocols and products optimized for FFPE-DNA, to limit additional damage to DNA during a process of extraction. - Performing DNA quality control before library preparation
DNA quality control should include assessing the yield and analyzing the fragmentation state26. The average fragment length correlates with coverage uniformity and is one of the most important quality criteria in FFPE-DNA sequencing. It can be performed using PCR or microfluidics capillary electrophoresis of DNA to visualize fragmentation patterns and compare sizing (and quantity) to standards. - Enzymatic repair of damaged DNA
The number of unique reads can be increased using a BER-based protocol27(Base Excision Repair), where enzymes, such as DNA glycosylases and endonucleases remove damaged bases. - Library preparation specific for FFPE-DNA
When FFPE-DNA is prepared for sequencing, workflow adaptations can improve the outcome. Useful adaptations include higher DNA input amounts (if possible), mild shearing conditions, FFPE-specific kits, the use of UMIs, and replicate strategies. - Sequencing
In general, deeper sequencing is needed for FFPE-DNA than for undamaged fresh frozen DNA.
RNA-Seq on FFPE samples
Transcriptome profiling provides information of great value in clinical decision-making and clinical and academic research. FFPE samples are a treasure chest of molecular information, and RNA-Seq from FFPE-RNAs can offer critical insights into the molecular secrets of diseases. The main challenge with using FFPE-RNA for transcriptome profiling is the overall low quality of FFPE-RNA. This issue was unsolvable in the past, and due to technical constraints, most of the knowledge hidden in FFPE samples was unavailable. Luckily, nowadays, there are many workarounds and optimized protocols and products specifically developed for extracting RNA from FFPE samples and library preparation of FFPE-RNA, as well as a lot of knowledge on how to approach the RNA-Seq data analysis stemming from FFPE samples. This is demonstrated in a large-scale whole transcriptome profiling study8 focused on examining any intrinsic biases in the FFPE-derived gene expression data and assessing its utility in clinical applications. This study used over 3000 FFPE samples, and the authors have performed analysis of various types of replicates at every stage of RNA sample fixation/storage, extraction, and sequencing. They could show that “when quality is controlled, gene expression quantification of FFPE-derived RNA is robust, consistent and reproducible”.
Based on years of experience with transcriptome sequencing from FFPE samples, we are working on preparing a form of a FFPE RNA-Seq cheat sheet that will contain details on all the challenges, possible solutions, and best practices, including an experimental checklist you can take with you to the lab. Stay tuned! In the meantime, let’s look at some examples from research on what discoveries are possible from transcriptome profiling of FFPE samples.
For any given cancer, only a small fraction of the total cases has enough fresh tissue available for unbiased RNA expression studies. Furthermore, even fewer cases are associated with long term outcomes data, which is very relevant for e.g., breast cancer. Namely, estrogen receptor (ER)+ breast cancer has a 5-year survival rate of greater than 95 % but as high as 40 % progression to metastasis in 10–20 years. One approach to address the barriers of “limited outcomes data” in cancer research is to use FFPE tissue for RNA sequencing. By optimizing a pipeline28 specifically for FFPE RNA-Seq, authors could use archival FFPE breast cancer tissues to distinguish between ER+ and ER- breast cancer cases. They compared FFPE data to publicly available databases obtained from fresh tissues to confirm gene expression fidelity. Another study used RNA-Seq on FFPE samples of breast cancer patients to rediscover RNA biomarkers for disease recurrence risk previously identified by RT-PCR29.
MicroRNAs (miRNAs) have emerged as biomarkers and predictive tools and as therapeutic targets in cancer. Focusing on clear cell renal cell carcinoma (ccRCC), a subtype of kidney cancer, researchers wanted to obtain a miRNA profile from cancer tissue samples using small RNA-Seq (in this case miRNA-Seq). They were interested in seeing if they could obtain good-quality profiles from readily available FFPE samples, and they reported that FFPE samples are a viable source of miRNA-sequencing material, comparable to fresh tissue samples30.
Future of NGS using FFPE samples
FFPE tissue samples are set to play a major role in the future of personalized healthcare. Large datasets that target cancer, like The Cancer Genome Atlas and the International Cancer Genome Consortium provide a large amount of genetic information on different cancers. Improvements in extracting usable material from FFPE samples and analyzing the resultant data mean that a huge archive of samples will be added to the global pool of biospecimens for analysis.
Experiments from Lexogen’s bench: FFPE RNA-Seq
Differential gene expression analysis is one of the most common RNA-Seq analyses from FFPE samples. In cancer research, this type of analysis is very often performed to compare gene expression profiles between healthy and tumor tissue. We have conducted a 3’ mRNA-Seq experiment on RNA isolated from human kidney tumor FFPE tissue samples and matched normal, healthy FFPE tissue. Reads were mapped against the human genome (hsa_GRCh38.94), and gene ontology enrichment and pathway analysis were done, focusing on different cancer “marker” genes31, such as various oncogenes (e.g. KRAS, MYC), or genes promoting cancer-specific metabolism32 (such as dependency on oxidative phosphorylation – OXPHOS). In total~16,950 genes were detected. Indeed, we could see that pathways associated with cancer proliferation and cancer-specific metabolism are upregulated. In contrast, genes driving apoptosis and immune system defence responses were downregulated in tumor FFPE samples vs. healthy FFPE samples (Figure 3).
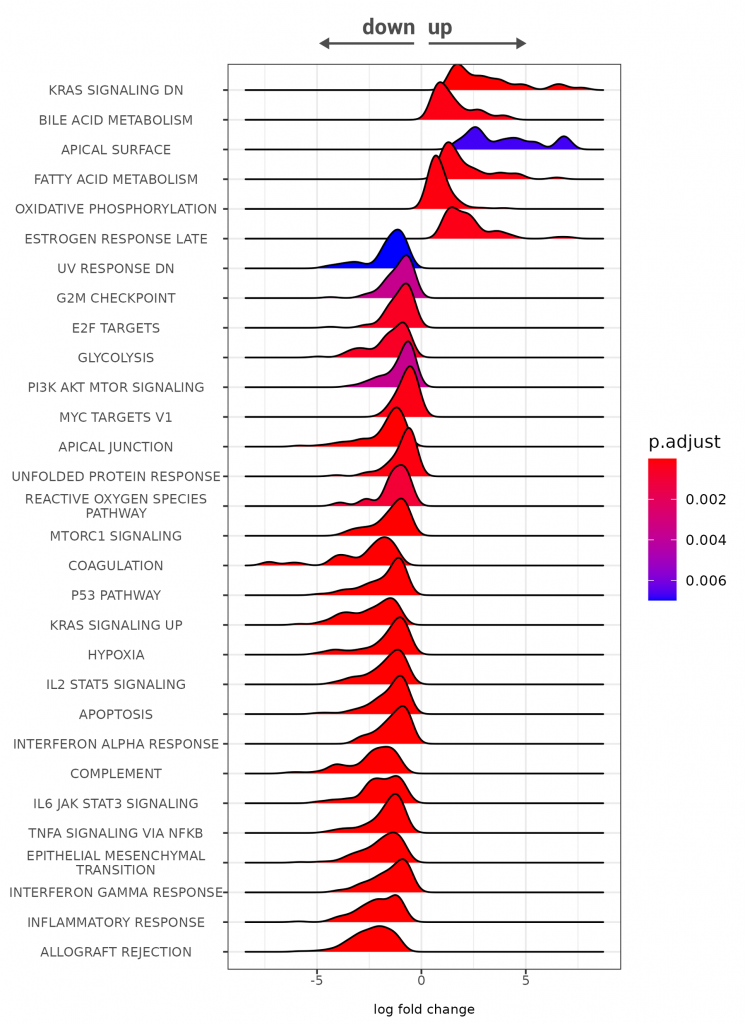
Figure 3 | Gene ontology enrichment analysis of human kidney tumor FFPE vs. (matched) normal FFPE tissue. RNA was extracted from FFPE samples with Split One-Step FFPE RNA Extraction, and 20 ng total RNA was used for 3’mRNA-Seq library preparation using QuantSeq FFPE kit. FFPE RNA quality: Human Kidney Tumor: DV200 = 45 %, Human Kidney Normal: DV200 = 50 % (see NOTE1). Sequencing was done with 3 M reads/sample and mapped against the human genome (hsa_GRCh38.94). Differential gene expression was performed with DESeq2. GESA plot for pathway enrichment was constructed using a threshold of padj* (see NOTE 2) <0.01.
NOTE 1: How is RNA quality measured for FFPE samples?
RNA quality is routinely measured using microfluidics devices. Instruments, such as Bioanalyzer or Fragment Analyzer provide traces of the RNA length profile, as well as an assessment of the RNA concentration and quality. High quality RNA categorized on a numerical scale from 1 – 10, called RIN (RNA Integrity Number, typically provided by Bioanalyzer) or RQN (RNA Quality Number, typically provided by Fragment Analyzer). The regions assessed to measure this score are for example the peaks of the rRNA for 18S and 23S in mammals. While the measurement differs slightly between instruments, the values are comparable. The higher the score (the closer to 10), the higher the RNA quality. However, FFPE RNA is often so degraded that rRNA peaks are no longer discernible and RIN or RQN scores cannot be determined or are not informative. Therefore, degraded and FFPE RNA quality is assessed by the so called DV200 value. DV200 values are a measure of the size distribution of RNA in a sample and directly state the fraction of RNA that is longer than 200 nt. In general, DV200 values can vary strongly depending on the preparation, storage, age, and extraction method for the sample. DV200 values <30% are considered poor quality by many RNA-Seq kit providers and caution should be applied when proceeding with samples below this threshold. Higher input RNA amounts may be required for library preparation, or samples might not be useable for some applications, e.g., target enrichment.
NOTE 2: How is significance determined for differential expression?
The adjusted p-value (padj) is more stringent as the p-value (or significance score under the null hypothesis). The padj also controls for the false discovery rate via the Benjamini-Hochberg method, whereby the stringency is increased the lower the threshold for padj is, i.e., the lower the likelihood of false discovery. The threshold for padj in this experiment is padj < 0.01 (highly stringent). For more information, see RNA Lexicon Chaper 13.
3’ mRNA-Seq is perfect for gene expression profiling, and we will talk more about that in one of our future blog posts. However, if we want to look at the whole transcriptome, we need to use a different approach—whole transcriptome sequencing (WTS). WTS allows a deeper look into the transcriptome and gives information about the entire transcript body and non-coding transcripts, e.g., long non-coding (lnc)RNAs, identified as potent biomarkers connected to several diseases.
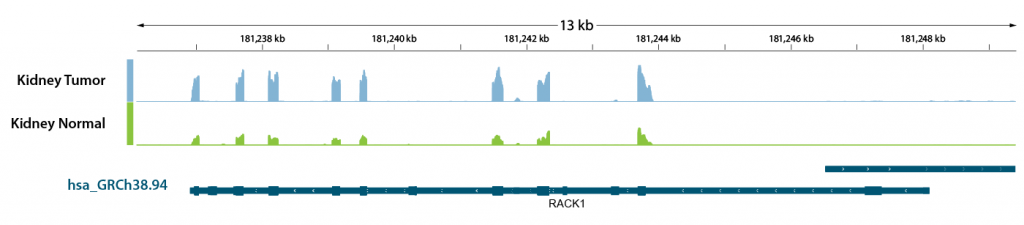
We used the same RNA isolated from human kidney tumor FFPE tissue samples and matched normal, healthy FFPE tissue, but we performed a WTS experiment this time. As a showcase, we looked at RACK1 (Receptor for Activated C Kinase 1), located at the 5q35.3 gene, implicated in 5q copy number gains and upregulation in kidney cancer and associated with the interferon-alpha inflammatory response33. As shown on Figure 4, we can see the upregulation of RACK1 in kidney tumor FFPE samples. WTS also allows us to see which RACK1 exons are being expressed – observe the “spikes” in the upper panel (Kidney Tumor, blue) and middle panel (Kidney Normal, green), and the dark blue blocks in the hsa_GRCh38.94 gene body representing exons in the lowest panel.
Figure 4 | Rack1 transcript body coverage in human kidney tumor FFPE vs. matched normal FFPE tissue. RNA was extracted with Split One-Step FFPE RNA Extraction from FFPE samples, and 20 ng total RNA was used for whole transcriptome sequencing with library preparation kit CORALL FFPE. FFPE RNA quality: Human Kidney Tumor: DV200 = 45%, Human Kidney Normal: DV200 = 50%. Reads were mapped against the human genome (hsa_GRCh38.94) and coverages were analyzed using genome viewer.
DNA-Seq and RNA-Seq Service from FFPE samples
As we learned in this blog, FFPE samples are a precious source of molecular information and applicable in many branches of modern research. However, they are challenging to work with, requiring more expertise, optimizations, and often more time and resources than sample types where nucleic acids are of higher quality, such as fresh, or fresh frozen samples. We at Lexogen have a team of NGS experts with many years of experience of performing DNA-Seq and RNA-Seq (both gene expression profiling and whole transcriptome sequencing) from FFPE samples. Our services options are versatile, and include everything from initial consultations, over DNA and RNA extraction, library preparation and sequencing, all up to NGS data analysis – and are fully customizable to fit the requirements of your projects.
References
- Gaffney EF, Riegman PH, Grizzle WE, Watson PH. Factors that drive the increasing use of FFPE tissue in basic and translational cancer research. Biotech Histochem. 2018;93(5):373-386. doi:10.1080/10520295.2018.1446101
- Paluch BE, Glenn ST, Conroy JM, et al. Robust detection of immune transcripts in FFPE samples using targeted RNA sequencing. Oncotarget. 2017;8(2):3197-3205. doi:10.18632/oncotarget.13691
- Wang D, Scheuenpflug J, Feng Z. 84 Comparative analysis of ImmunoID NeXT™ and ACE ImmunoID™ next generation sequencing platforms for investigating tumor-immune interactions to enable precision oncology driven biomarker discovery. J Immunother Cancer. 2021;9(Suppl 2):A92-A92. doi:10.1136/jitc-2021-SITC2021.084
- Martínez-Hernández R, Marazuela M. MicroRNAs in autoimmune thyroid diseases and their role as biomarkers. Best Pract Res Clin Endocrinol Metab. 2023;37(2):101741. doi:10.1016/j.beem.2023.101741
- Robetorye RS, Maguire A, Rosenthal AC, Rimsza LM. Profiling of lymphoma from formalin-fixed paraffin-embedded tissue. Semin Hematol. 2019;56(1):46-51. doi:10.1053/j.seminhematol.2018.05.003
- Keene CD, Wilson AM, Kilgore MD, Bruner LT, Postupna NO, Darvas M. Luminex-based quantification of Alzheimer’s disease neuropathologic change in formalin-fixed post-mortem human brain tissue. Lab Invest. 2019;99(7):1056-1067. doi:10.1038/s41374-018-0165-x
- Gao XH, Li J, Gong HF, et al. Comparison of Fresh Frozen Tissue With Formalin-Fixed Paraffin-Embedded Tissue for Mutation Analysis Using a Multi-Gene Panel in Patients With Colorectal Cancer. Front Oncol. 2020;10:310. doi:10.3389/fonc.2020.00310
- Newton Y, Sedgewick AJ, Cisneros L, et al. Large scale, robust, and accurate whole transcriptome profiling from clinical formalin-fixed paraffin-embedded samples. Sci Rep. 2020;10(1):17597. doi:10.1038/s41598-020-74483-1
- Xu Z, Zhang T, Chen H, et al. High-throughput single nucleus total RNA sequencing of formalin-fixed paraffin-embedded tissues by snRandom-seq. Nat Commun. 2023;14(1):2734. doi:10.1038/s41467-023-38409-5
- Mosele F, Remon J, Mateo J, et al. Recommendations for the use of next-generation sequencing (NGS) for patients with metastatic cancers: a report from the ESMO Precision Medicine Working Group. Ann Oncol. 2020;31(11):1491-1505. doi:10.1016/j.annonc.2020.07.014
- Cardoso F, van’t Veer LJ, Bogaerts J, et al. 70-Gene Signature as an Aid to Treatment Decisions in Early-Stage Breast Cancer. N Engl J Med. 2016;375(8):717-729. doi:10.1056/NEJMoa1602253
- Steiert TA, Parra G, Gut M, et al. A critical spotlight on the paradigms of FFPE-DNA sequencing. Nucleic Acids Research. 2023;51(14):7143-7162. doi:10.1093/nar/gkad519
- Moorcraft SY, Gonzalez D, Walker BA. Understanding next generation sequencing in oncology: A guide for oncologists. Crit Rev Oncol Hematol. 2015;96(3):463-474. doi:10.1016/j.critrevonc.2015.06.007
- Cappello F, Angerilli V, Munari G, et al. FFPE-Based NGS Approaches into Clinical Practice: The Limits of Glory from a Pathologist Viewpoint. J Pers Med. 2022;12(5). doi:10.3390/jpm12050750
- Solassol J, Ramos J, Crapez E, et al. KRAS mutation detection in paired frozen and Formalin-Fixed Paraffin-Embedded (FFPE) colorectal cancer tissues. Int J Mol Sci. 2011;12(5):3191-3204. doi:10.3390/ijms12053191
- Kerick M, Isau M, Timmermann B, et al. Targeted high throughput sequencing in clinical cancer settings: formaldehyde fixed-paraffin embedded (FFPE) tumor tissues, input amount and tumor heterogeneity. BMC Med Genomics. 2011;4:68. doi:10.1186/1755-8794-4-68
- Taylor JC, Martin HC, Lise S, et al. Factors influencing success of clinical genome sequencing across a broad spectrum of disorders. Nat Genet. 2015;47(7):717-726. doi:10.1038/ng.3304
- Robbe P, Popitsch N, Knight SJL, et al. Clinical whole-genome sequencing from routine formalin-fixed, paraffin-embedded specimens: pilot study for the 100,000 Genomes Project. Genetics in Medicine. 2018;20(10):1196-1205. doi:10.1038/gim.2017.241
- Astolfi A, Urbini M, Indio V, et al. Whole exome sequencing (WES) on formalin-fixed, paraffin-embedded (FFPE) tumor tissue in gastrointestinal stromal tumors (GIST). BMC Genomics. 2015;16:892. doi:10.1186/s12864-015-1982-6
- Feldman MY. Reactions of nucleic acids and nucleoproteins with formaldehyde. Prog Nucleic Acid Res Mol Biol. 1973;13:1-49. doi:10.1016/s0079-6603(08)60099-9
- ALY. PII: S0027-5107(97)00314-X.
- Sikorsky JA, Primerano DA, Fenger TW, Denvir J. DNA damage reduces Taq DNA polymerase fidelity and PCR amplification efficiency. Biochem Biophys Res Commun. 2007;355(2):431-437. doi:10.1016/j.bbrc.2007.01.169
- Chen G, Mosier S, Gocke CD, Lin M-T, Eshleman JR. Cytosine deamination is a major cause of baseline noise in next-generation sequencing. Mol Diagn Ther. 2014;18(5):587-593. doi:10.1007/s40291-014-0115-2
- Beland FA, Fullerton NF, Heflich RH. Rapid isolation, hydrolysis and chromatography of formaldehyde-modified DNA. J Chromatogr. 1984;308:121-131. doi:10.1016/0378-4347(84)80202-9
- Gilbert MTP, Haselkorn T, Bunce M, et al. The isolation of nucleic acids from fixed, paraffin-embedded tissues-which methods are useful when? PLoS One. 2007;2(6):e537. doi:10.1371/journal.pone.0000537
- Ottestad AL, Emdal EF, Grønberg BH, Halvorsen TO, Dai HY. Fragmentation assessment of FFPE DNA helps in evaluating NGS library complexity and interpretation of NGS results. Exp Mol Pathol. 2022;126:104771. doi:10.1016/j.yexmp.2022.104771
- Krokan HE, Bjørås M. Base excision repair. Cold Spring Harb Perspect Biol. 2013;5(4):a012583. doi:10.1101/cshperspect.a012583
- Pennock ND, Jindal S, Horton W, et al. RNA-seq from archival FFPE breast cancer samples: molecular pathway fidelity and novel discovery. BMC Med Genomics. 2019;12(1):195. doi:10.1186/s12920-019-0643-z
- Sinicropi D, Qu K, Collin F, et al. Whole transcriptome RNA-Seq analysis of breast cancer recurrence risk using formalin-fixed paraffin-embedded tumor tissue. PLoS One. 2012;7(7):e40092. doi:10.1371/journal.pone.0040092
- Strauss P, Marti H-P, Beisland C, et al. Expanding the Utilization of Formalin-Fixed, Paraffin-Embedded Archives: Feasibility of miR-Seq for Disease Exploration and Biomarker Development from Biopsies with Clear Cell Renal Cell Carcinoma. IJMS. 2018;19(3). doi:10.3390/ijms19030803
- Wu Y, Terekhanova NV, Caravan W, et al. Epigenetic and transcriptomic characterization reveals progression markers and essential pathways in clear cell renal cell carcinoma. Nat Commun. 2023;14(1):1681. doi:10.1038/s41467-023-37211-7
- DeBerardinis RJ, Chandel NS. Fundamentals of cancer metabolism. Sci Adv. 2016;2(5):e1600200. doi:10.1126/sciadv.1600200
- Li J-J, Xie D. RACK1, a versatile hub in cancer. Oncogene. 2015;34(15):1890-1898. doi:10.1038/onc.2014.127
Written by Masa Ivin, PhD
Photography by Mag. Amra Dedic