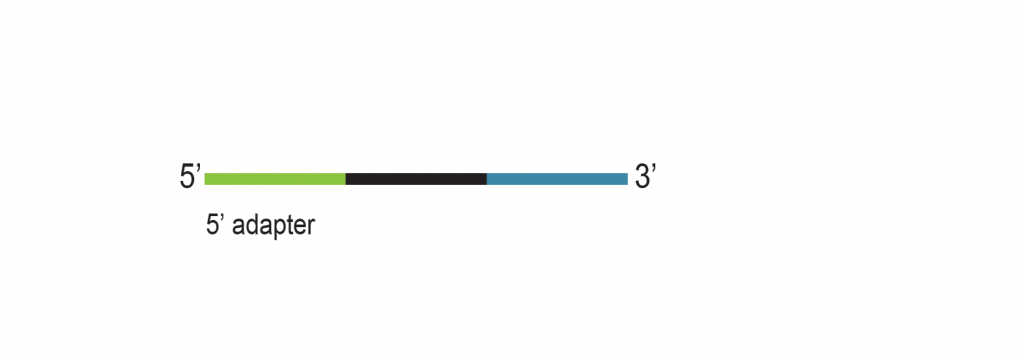miRVEL Discovery Small RNA-Seq Library Prep
miRVEL Discovery Small RNA-Seq Library Prep
miRVEL Discovery Small RNA-Seq Library Prep Kit provides cutting-edge technology for sRNA-Seq, specifically optimized for low input biofluids, such as blood and plasma samples, making it a perfect solution for circulating biomarker studies. The miRVEL Discovery technology utilizes modified oligonucleotides that effectively inhibit the formation of adapter dimers and eliminate the need for gel-size selection, thereby reducing hands-on time and streamlining the overall workflow.
Benefits

Unlock biofluid samples
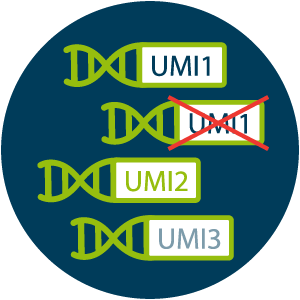
Precise quantification with UMIs
10 nt UDIs
Performance
miRVEL Discovery Small RNA-Seq Library Prep provides a cutting-edge solution for sRNA-Seq with exceptional performance on low input biofluid samples.
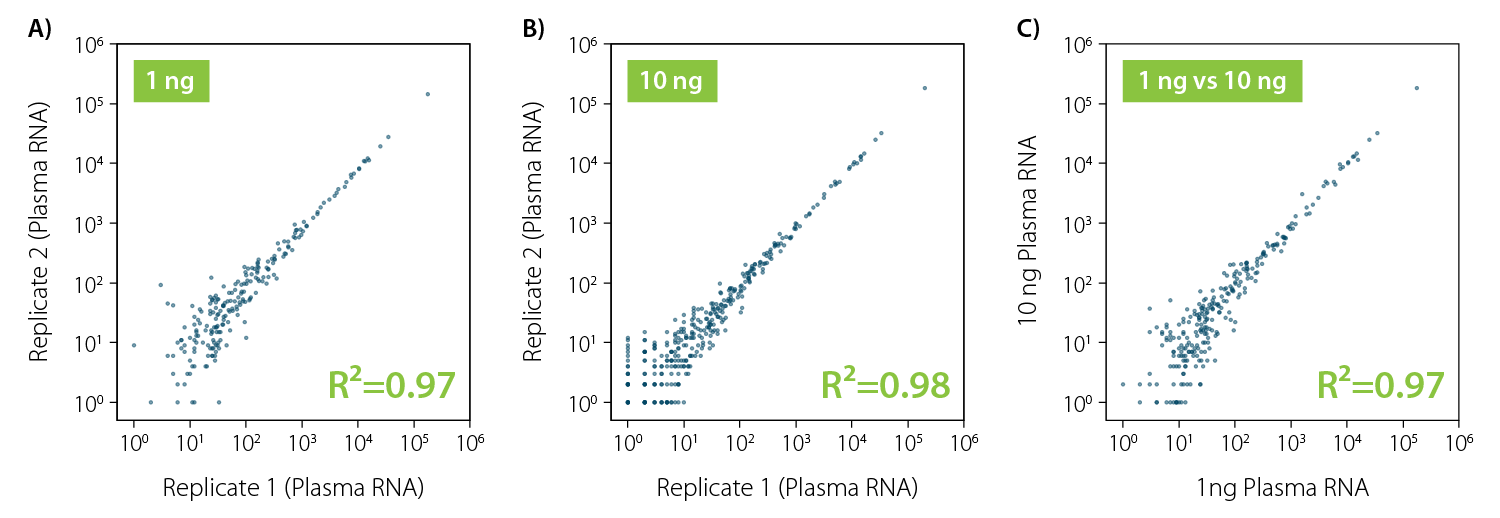
Outstanding Reproducibility and Correlation Across Different Concentrations
miRVEL Discovery Small RNA-Seq Library Prep generates excellent gene count correlation between replicates (Fig. 1A, 1B) even with low inputs (1 ng plasma RNA, Fig. 1A). Moreover, the performance remains consistent across a range of inputs (Fig. 1C), which highlights the robustness of the miRVEL Discovery protocol.
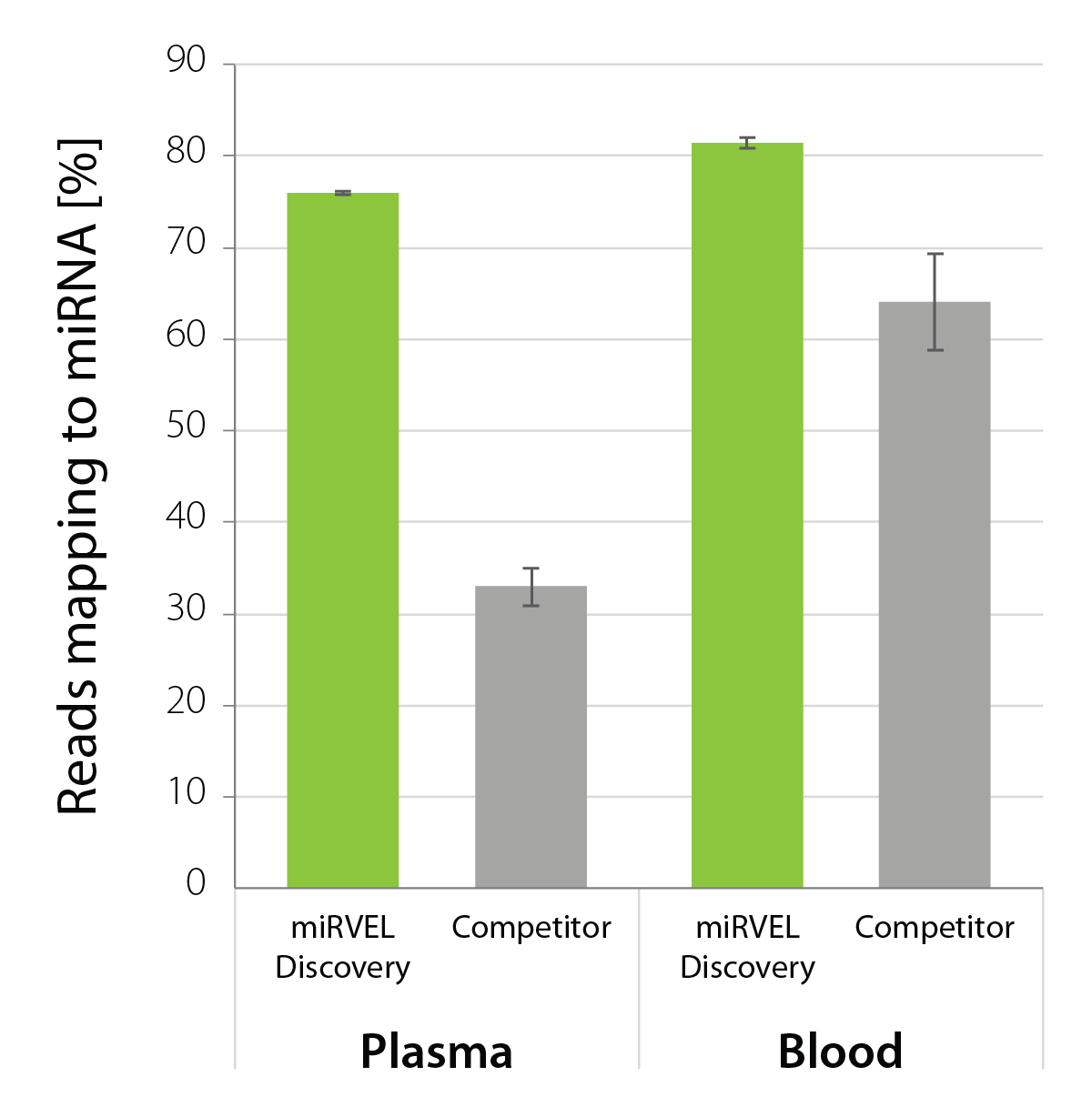
Exceptional Rate of Reads Mapping in miRNAs for Biofluid Samples
miRVEL Discovery Small RNA-Seq Library Prep exhibits an exceptional rate of reads mapping to miRNA for libraries generated from 10 ng of human plasma RNA and 1 ng of human blood RNA (Fig. 2).
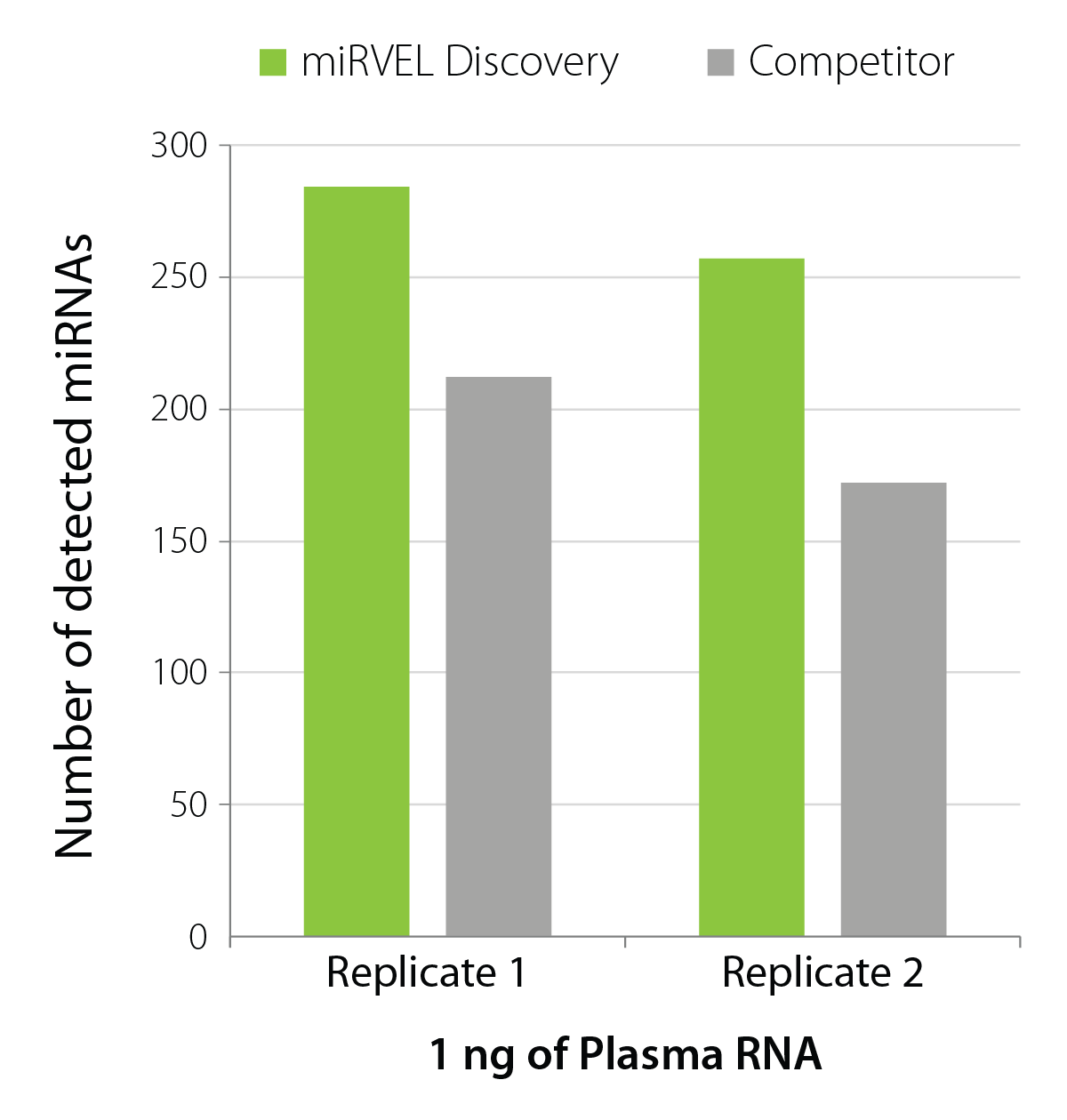
Excellent miRNA Detection in Low-input Biofluid Samples
miRVEL Discovery Small RNA-Seq Library Prep demonstrates a high detection of miRNAs in low-input plasma samples (Fig. 3), making it an outstanding solution for biomarker research.
Workflow
The miRVEL Discovery Small RNA-Seq Library Prep Kit features a streamlined workflow that allows the entire library preparation process to be completed in under 7 hours. The methodology utilizes modified oligonucleotides that effectively prevent adapter dimer formation, eliminating the need for time-consuming gel-size selection.
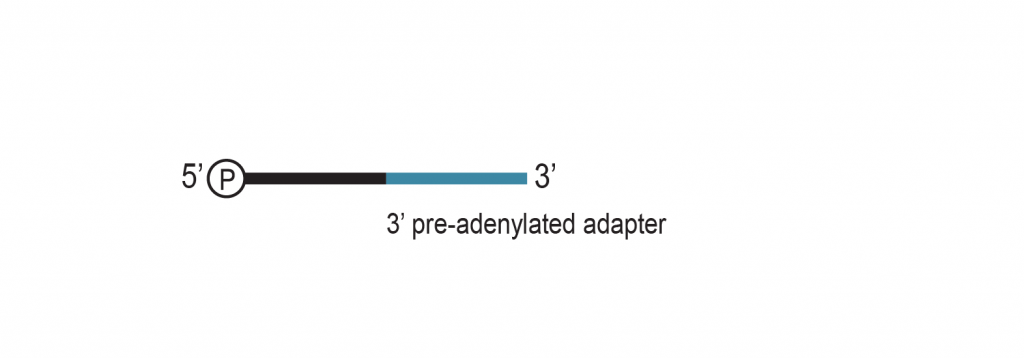
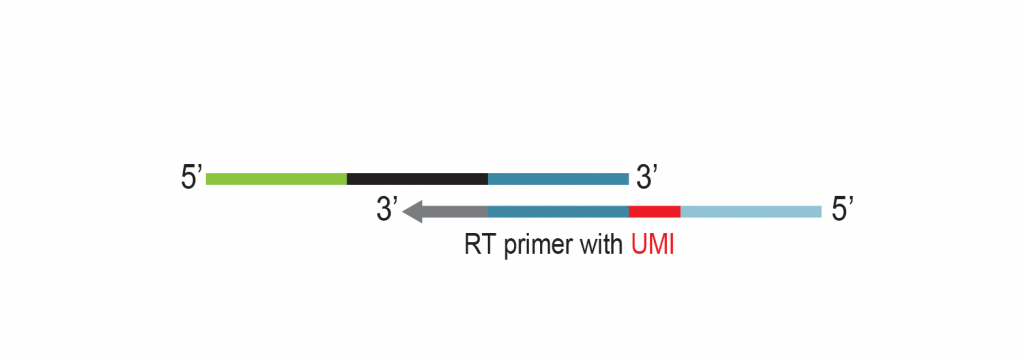
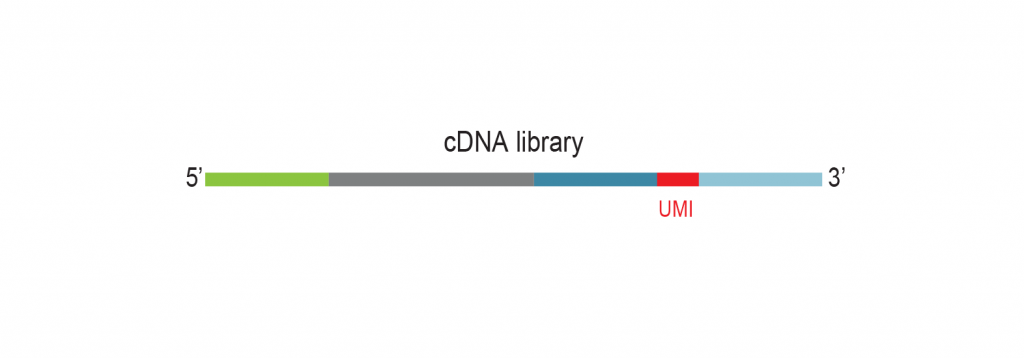
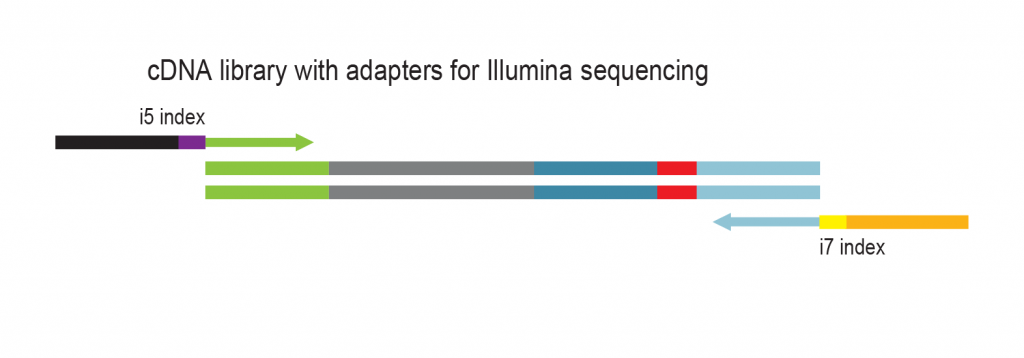
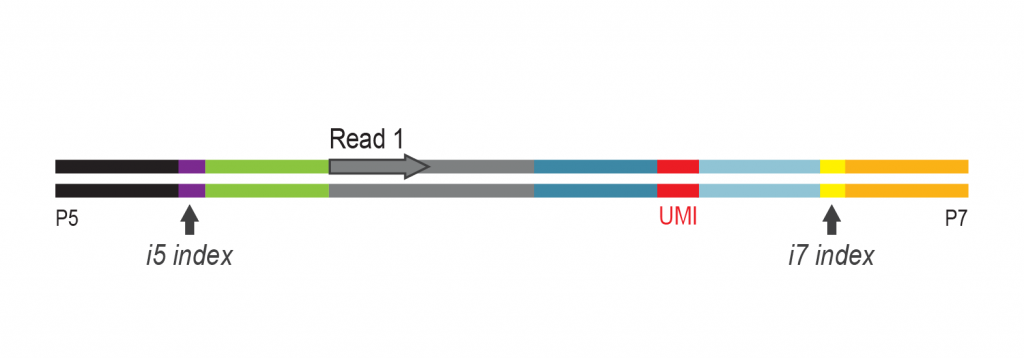
The input RNA, flanked by 5’ and 3’ adapters, is converted into cDNA using an RT primer that binds to the 3’ adapter. This RT primer includes a unique molecular identifier (UMI) that is incorporated into each cDNA molecule. Additionally, the RT primer introduces a universal sequence that is recognized by the sample indexing primers during library amplification.
During PCR library amplification, unique dual i7 and i5 indices, along with complete adapter sequences, are incorporated. The amplified libraries undergo a final bead-based purification.
FAQ
Frequently Asked Questions
Access our frequently asked question (FAQ) resources via the buttons below.
Please also check our General Guidelines and FAQ resources!
How do you like the new online FAQ resource? Please share your feedback with us!
Downloads
Safety Data Sheet
If you need more information about our products, please contact us through support@lexogen.com or directly under +43 1 345 1212-41.
Ordering Information
| Cat. No. | Product Name |
| 242.24 | miRVEL Discovery Small RNA-Seq Library Prep Kit, 24 preps |
| 242.96 | miRVEL Discovery Small RNA-Seq Library Prep Kit, 96 preps |
Buy from our Webstore
Need a web quote?
You can generate a web quote by Register or Login to your account. In the account settings please fill in your billing and shipping address. Add products to your cart, view cart and click the “Generate Quote” button. A quote in PDF format will be generated and ready to download. You can use this PDF document to place an order by sending it directly to sales@lexogen.com.
Web quoting is not available for countries served by our distributors. Please contact your local distributor for a quote.