We are happy to share news about the paper “cDNA Library Enrichment of Full Length Transcripts for SMRT Long Read Sequencing” published last week by a group from Max Planck Institute for Plant Breeding Research in Cologne (Germany). In this study they wanted to utilize the power of long read sequencing provided by Pacific Bioscience Iso-Seq technology for a thorough study of transcription start (TSS) and end sites (TES) in the well annotated Arabidopsis Thalia genome and the complex polyploid genome of Triticum aestivum. The group has compared the TeloPrime Full-Length cDNA Synthesis protocol from Lexogen with the most commonly used SMARTer PCR cDNA Synthesis Kit for preparing Full-Length cDNA for Iso-Seq library prep.
The authors reported that one major finding is that TeloPrime – in contrast to SMARTer – is able to distinguish between full-length and truncated transcripts. Therefore full-length transcripts are not appropriately represented in the sequencing data if SMARTer technology is used. The differences between the two technologies are explained in detail in the paper.
In the first part of the study experiments with Arabidopsis thaliana samples prepared with TeloPrime yielded substantially higher number of High Quality reads aligned to the annotated TSS than in samples prepared with SMARTer. TeloPrime samples also had more reads mapping upstream of annotated TSS, showing its potential for improvement of the TSS predictions even for very well annotated genomes (Figure below).
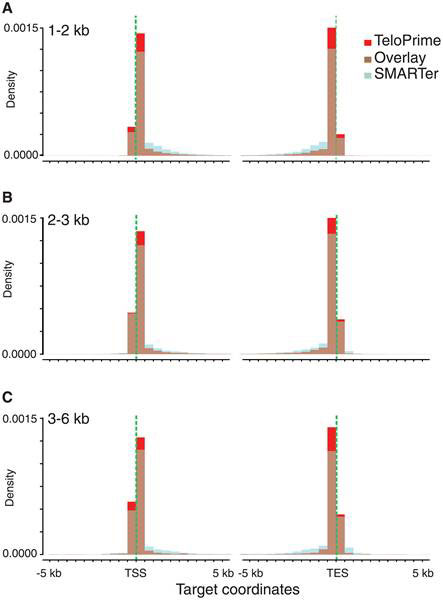
Figure. Transcription start and end site enrichment using the TeloPrime Full-Length cDNA Amplification kit (Lexogen). (A-C) Superimposed density plots of the PacBio Iso-Seq alignment coordinates against the 5’ and 3’ ends of their targets. TeloPrime cDNA libraries (red bars), SMARTer cDNA libraries (light blue bars), overlay of the two protocols (brown bars). (A) 1–2 kb size fraction; (B) 2–3 kb size fraction; (C) 3–6 kb size fraction. 10 kb around the annotated gene start and end coordinates are shown on the x-axis. Vertical green dashed lines highlight annotated target start and end sites.
In the second part sequencing of Triticum aestivum transcriptome using TeloPrime-based PacBio Iso-Seq showed a high potential for the improvement of the current wheat annotation. The authors confirmed that TeloPrime can be successfully used for the annotation of gene models and the identification of novel transcription sites, gene homologs, splicing isoforms and previously unidentified gene loci.
Read the complete paper: http://journals.plos.org/plosone/article?id=10.1371%2Fjournal.pone.0157779
Learn more about TeloPrime Full-Length cDNA Amplification Kit.
Contact us if you have any questions at info@lexogen.com.








