Do you have any questions?
Is LUTHOR HD the right solution for your RNA research? Let’s find out!
LUTHOR HD POOL
Down to single cells or 10 pg RNA input
Down to 2 EUR per library
Up to 36,864 samples per run
Up to 2,548 saved pipet tips (96 preps)
LUTHOR High-Definition Pool Single-Cell 3′ mRNA-Seq Library Prep Kit
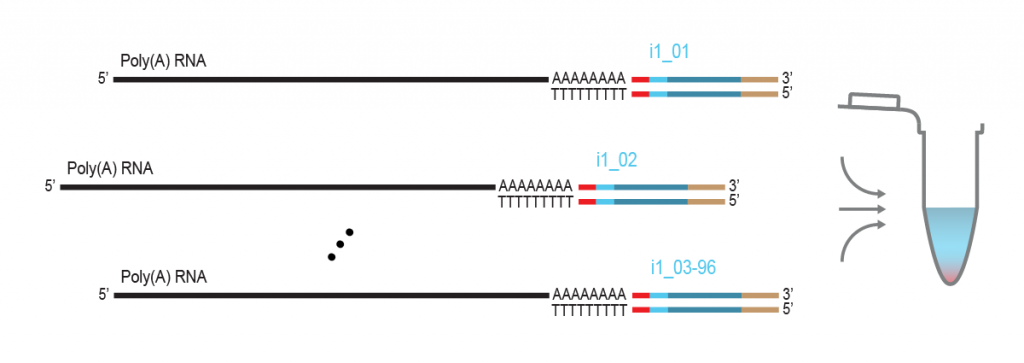
LUTHOR HD Pool benefits from the original THOR technology to amplify minute amounts of input RNA. The Pool version includes carefully selected LPi1 sample barcodes, which are added at the very start of the library preparation. Once LPi1 barcodes are linked to each sample, pooling of 8 – 96 samples in one tube simplifies the protocol significantly reducing hands-on time and consumable costs. LUTHOR HD Pool is ideal for experiments with homogeneous sample types.
Running large scale projects?
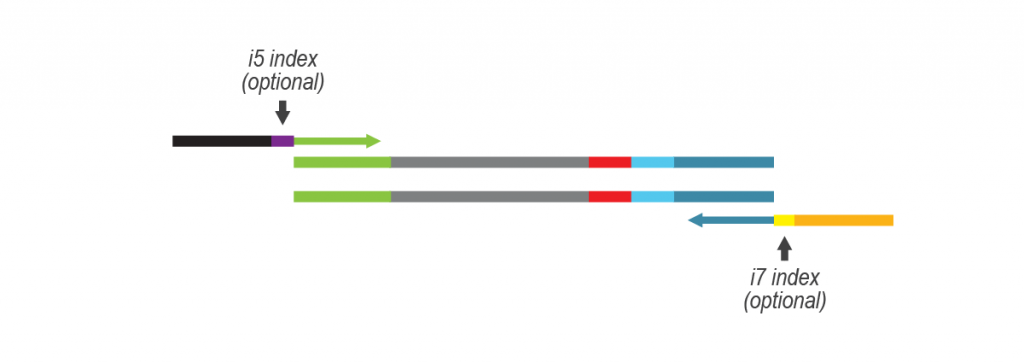
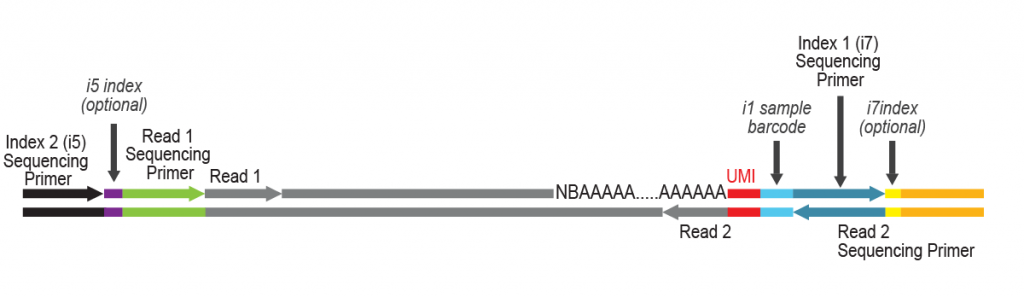
For more samples, we recommend adding Lexogen’s patented 12 nt UDI for triple indexing (LPi1, i5, i7), and up to 36,864 different barcode combinations. Lexogen UDI Sets are available in 96-well plates and allow mixing of different Lexogen libraries (e.g., CORALL, QuantSeq, LUTHOR) with LUTHOR HD Pool libraries in sequencing lanes, contact support@lexogen.com for more information.
Scaling up? Large project? Benefit from our bulk configurations and dynamic pricing schemes! Contact us at sales@lexogen.com for a personalized quotation. When opting for bulk configurations, UDI are included in each kit.
RNA-Seq directly from lysates
LUTHOR HD Pool does not require RNA extraction. If you start from isolated cells, a dedicated lysis buffer is provided in the kit.
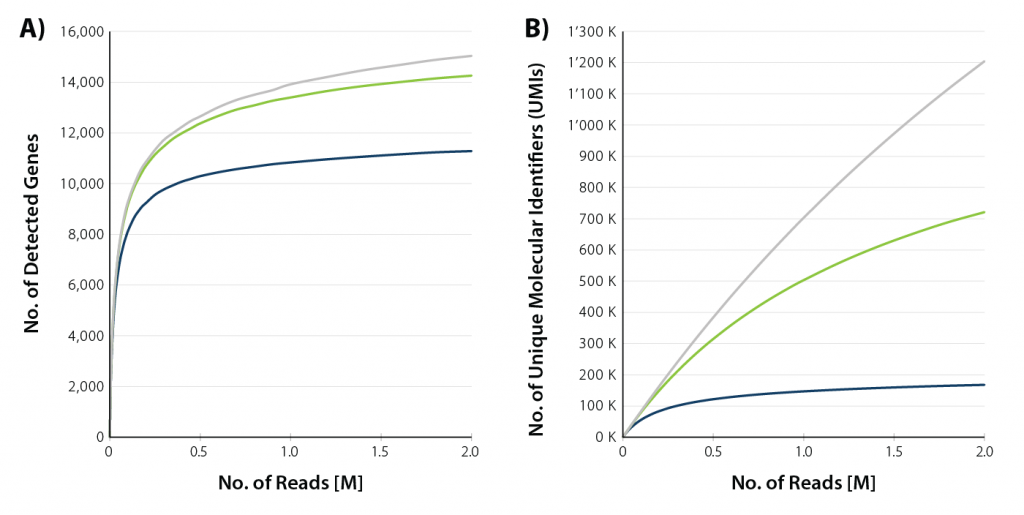
Sample input ranges from 10 pg (equivalent to the RNA content in one single cell) up to 10 ng. Read depth of 1 M reads / sample is usually sufficient to capture the full gene diversity, while 5 M reads / sample may be necessary for a comprehensive analysis of all transcripts (typically, there are multiple transcripts for each gene).
Add-on kits:
- If you want to combine more than 96 libraries at once, we recommend our 12 nt UDI Sets.
- If you prepare 7 or more pools from a single LUTHOR HD Pool Kit, you will require the PCR Add-on Kit for Illumina (Cat. No. 208)
Performance
High Definition in a Pool
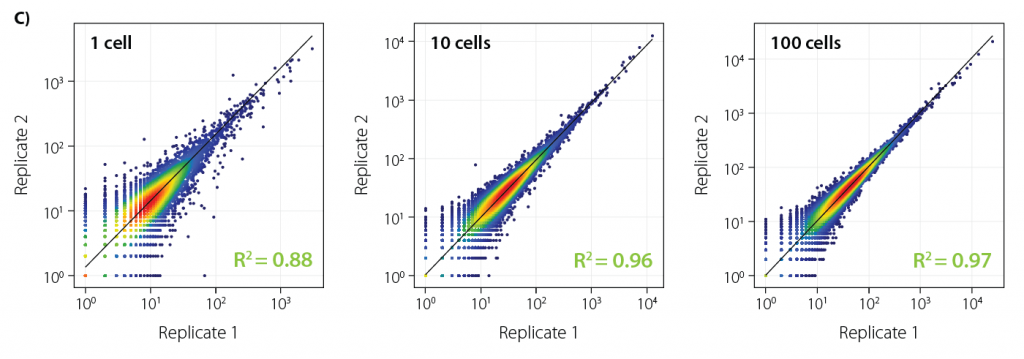
High Gene Detection Homogeneity between Samples
| A) | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| (cells) RSD: 1.6% | ||||||||||
| 86.3 | 85.7 | 88.2 | 86.6 | 85.4 | 85.1 | |||||
| 87.6 | 85.5 | 85.4 | 82.1 | 86.6 | 88.3 | |||||
| 82.1 | 86.6 | 88.3 | 86.1 | 87.2 | 87.8 | |||||
| 87.7 | 88.8 | 87.2 | 86.4 | 86.2 | 86.6 | |||||
| 86.2 | 86.1 | 88.4 | 88.5 | 85.8 | 85.2 | |||||
| 84.5 | 85.5 | 86.7 | 87.9 | 86.2 | 87.0 | |||||
| 85.4 | 87.3 | 87.0 | 86.9 | 86.4 | 86.8 | |||||
| 87.3 | 87.2 | 89.3 | 88.1 | 87.6 | 87.5 | |||||
| (UHRR) RSD: 1.4% | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| 80.6 | 81.4 | 80.9 | 80.8 | 80.1 | 80.7 | 79.9 | 77.7 | 80.6 | 80.0 | 80.8 | 81.5 |
| 79.9 | 81.3 | 81.3 | 81.1 | 81.5 | 80.6 | 80.8 | 80.8 | 80.6 | 81.3 | 80.0 | 79.6 |
| 81.0 | 80.7 | 80.7 | 80.2 | 79.8 | 80.5 | 80.8 | 75.5 | 80.3 | 79.1 | 80.4 | 81.3 |
| 80.5 | 81.6 | 80.7 | 80.6 | 79.1 | 81.0 | 80.7 | 81.8 | 81.3 | 80.6 | 79.6 | 79.6 |
| 80.0 | 80.7 | 81.0 | 79.6 | 80.8 | 79.2 | 80.9 | 78.0 | 81.4 | 81.0 | 80.6 | 81.0 |
| 82.3 | 80.4 | 80.9 | 77.0 | 80.5 | 81.4 | 82.2 | 80.9 | 80.1 | 81.7 | 80.3 | 81.2 |
| 80.9 | 80.7 | 80.6 | 81.1 | 77.3 | 81.5 | 81.6 | 80.2 | 79.7 | 80.0 | 80.0 | 79.6 |
| 79.9 | 82.0 | 80.3 | 80.8 | 79.8 | 80.5 | 81.8 | 79.1 | 79.9 | 78.3 | 78.7 | 80.9 |
| B) | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| (cells) RSD: 6.7% | ||||||||||
| 11’160 | 10’180 | 10’447 | 10’149 | 10’393 | 10’503 | |||||
| 10’880 | 9’970 | 9’878 | 7’697 | 11’257 | 10’942 | |||||
| 9’437 | 11’178 | 10’095 | 11’000 | 10’910 | 10’807 | |||||
| 10’793 | 9’607 | 11’016 | 9’758 | 10’225 | 10’550 | |||||
| 10’667 | 11’116 | 9’275 | 9’661 | 10’391 | 10’012 | |||||
| 11’676 | 10’557 | 10’967 | 11’438 | 10’126 | 10’073 | |||||
| 10’311 | 10’196 | 11’039 | 10’526 | 10’225 | 9’465 | |||||
| 10’515 | 10’531 | 10’644 | 11’934 | 10’523 | 10’446 | |||||
| (UHRR) RSD: 3.2% | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| 11956 | 12290 | 11935 | 12110 | 12197 | 12185 | 11639 | 10894 | 12123 | 11157 | 11963 | 12265 |
| 10976 | 12180 | 11932 | 12400 | 12461 | 12101 | 11724 | 11769 | 12219 | 12207 | 11885 | 10923 |
| 12495 | 12726 | 12152 | 11899 | 11925 | 11943 | 12531 | 12104 | 12281 | 12464 | 12370 | 12426 |
| 12090 | 12565 | 12283 | 12065 | 11839 | 11758 | 11987 | 12578 | 12016 | 12090 | 11974 | 12225 |
| 11604 | 12133 | 12103 | 12004 | 11818 | 11873 | 12178 | 11392 | 12158 | 12057 | 11896 | 12233 |
| 12816 | 11787 | 12238 | 11696 | 12403 | 12353 | 12171 | 11827 | 11552 | 12136 | 11991 | 12356 |
| 12134 | 11977 | 12463 | 12358 | 11681 | 12359 | 12849 | 12251 | 11902 | 10541 | 11750 | 11679 |
| 11918 | 11851 | 12390 | 12175 | 11995 | 11889 | 11827 | 11888 | 11972 | 11588 | 11652 | 12136 |
Minimal Cross-contamination
Pooled ngs library preparation approaches are inherently prone to some level of contamination between samples. At Lexogen, we made sure that cross-contamination is reduced to almost undetectable levels. Even when tips are not changed!
| A) | ||||||
|---|---|---|---|---|---|---|
| UHRR | Arabidopsis thaliana | Cross-contamination | ||||
| UHRR (6 samples) | 3,096,073 | 649 | 0.0% | |||
| Arabidopsis thaliana (2 samples) | 3,299 | 1,128,368 | 0.3% | |||
| total | 0.1% | |||||
| B) | ||||||
| UHRR | Arabidopsis thaliana | Cross-contamination | ||||
| UHRR (6 samples) | 3,245,024 | 595 | 0.0% | |||
| Arabidopsis thaliana (2 samples) | 3,208 | 1,175,775 | 0.3% | |||
| total | 0.1% | |||||
Table 2 | Assessment of Cross-contamination between Pooled Samples. 2 samples of Arabidopsis thaliana RNA (200 pg each) and 6 samples of UHRR (200 pg each) were pooled and prepared with LUTHOR HD Pool. Gene counts measured on data downsampled to 1M read/sample (read counts are cumulated in the table). Cross-contamination calculated as the percentage of cross-species gene counts over total species-specific gene counts.
A) Tips are changed between each sample. B) Tips are not changed between samples during THOR reaction.
Workflow
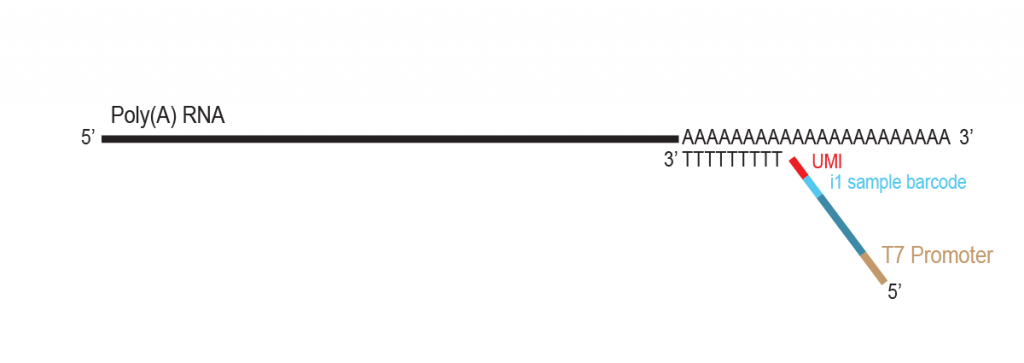
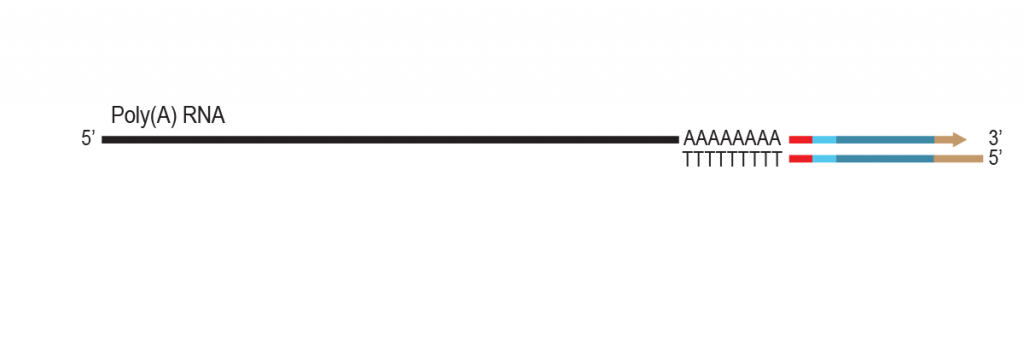
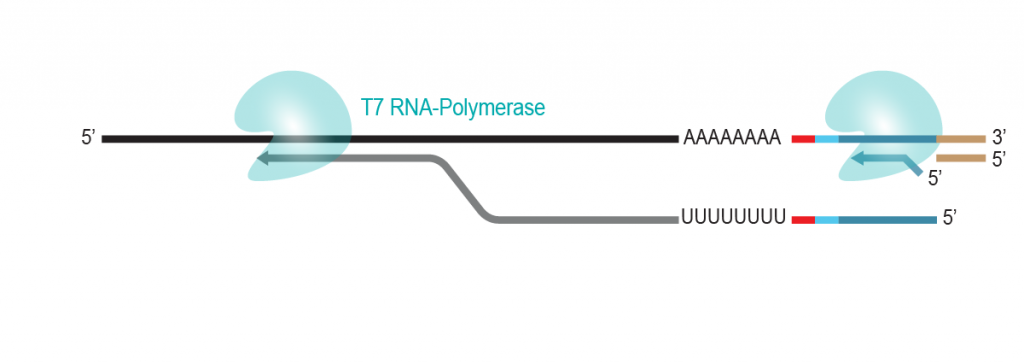
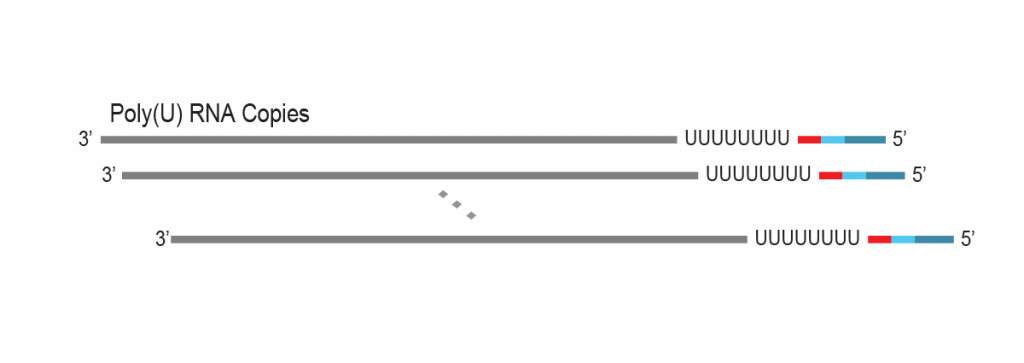
During a 2-hour (optional: overnight) in vitro transcription step the original mRNA template is copied repeatedly by linear amplification.
FAQ
Frequently Asked Questions
Access our frequently asked question (FAQ) resources via the buttons below.
Please also check our General Guidelines and FAQ resources!
How do you like the new online FAQ resource? Please share your feedback with us!
Downloads
LUTHOR HD Pool Single-Cell 3' mRNA-Seq
Safety Data Sheet
If you need more information about our products, please contact us through support@lexogen.com or directly under +43 1 345 1212-41.
Ordering Information
| Cat. No. | Product Name |
| 205.96 | LUTHOR High-Definition Pool Single-Cell 3′ mRNA-Seq Library Prep Kit, 96 preps |