Lexogen’s Data Analysis Solutions
Your analyzed data is just a few clicks away!
Based on state-of-the-art Lexogen’s proprietary pipeline
User-friendly – no bioinformatic skills required
Lexogen’s Data Analysis Solutions
We offer an automated solution for QuantSeq and CORALL data analysis based on state-of-the-art Lexogen’s proprietary pipeline. Thus, every user, even without bioinformatics experience, can analyze RNA-seq data conveniently and quickly.

QuantSeq 3′ mRNA-Seq
QuantSeq technology is a simple method focusing on 3’ ends of mRNA molecules to assess gene expression profiles by RNA sequencing easily. For more information, visit QuantSeq Webpage.
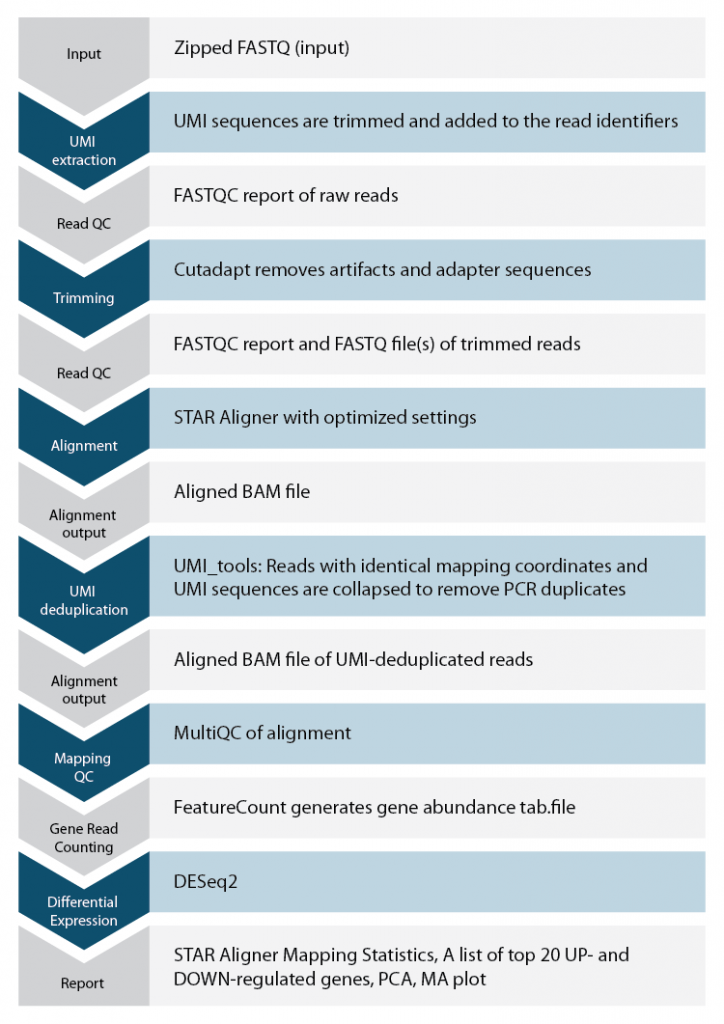
Lexogen’s automated solution for data analysis allows researchers to analyze QuantSeq samples conveniently and quickly, even for users without bioinformatics experience. The workflow for our QuantSeq pipeline is shown on the right.
Each QuantSeq FWD and REV kit is provided with codes for free data analysis on Lexogen’s data analysis solution, including differential expression. Data analysis codes for additional runs or bigger files can be purchased from Lexogen. Please get in touch with us at sales@lexogen.com
CORALL RNA-Seq
The CORALL Total RNA-Seq Library Prep Kit enables fast and cost-efficient generation of UMI labelled, stranded libraries for whole transcriptome analyses using Illumina® NGS platforms. For more information, visit CORALL Webpage.
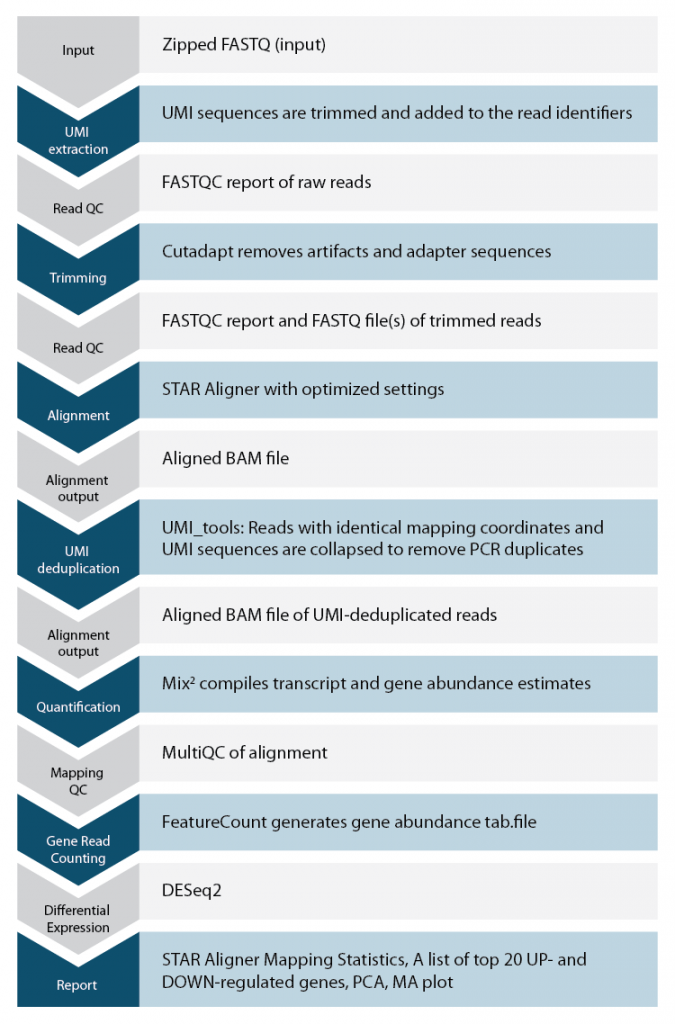
Lexogen’s automated solution for data analysis allows researchers, even those without bioinformatics experience, to analyze CORALL samples in a convenient and fast way. The workflow for our CORALL pipeline is shown on the right.
Data analysis codes for CORALL data analysis are available for purchase. Please contact us at sales@lexogen.com.
NOTE! Data Analysis on Lexogen’s data analysis solution is available for a various range of species. Reference genomes for new species can be added upon request. Please note this will incur a fee.
FAQ
Frequently Asked Questions
Access our frequently asked question (FAQ) resources via the buttons below.
Please also check our General Guidelines and FAQ resources!