
Let us help you to improve your SARS-CoV-2 sequencing workflows
- Reliable method to detect and trace viral variants
- Fast and easy ready-to-sequence library prep for Illumina
SARS-CoV-2 ARTIC Panel for Illumina
Lexogen’s SARS-CoV-2 ARTIC Panel for Illumina combines Illumina-compatible ARTIC primer pools with high-performance NGS reagents and indexing solutions.
Applications
Epidemiologic surveillance sequencing for variant identification and tracking.
Monitoring of viral evolution, spread, and transmission routes.
Features
Streamlined, fast protocol for SARS-Cov-2 whole genome sequencing.
Fast and easy ready-to-sequence library construction by indexing PCR.
Partial Illumina adaptors introduced during amplicon generation.
Compatible with Lexogen’s Unique Dual Indexing Solutions for multiplexing of 384 samples.
Overview

Figure 1 | Overview of the Lexogen ARTIC Workflow.
Description
The NGS-based panel allows to sequence the complete viral genome found in SARS-CoV-2 positive samples starting from cDNA. The virus material is enriched by amplicon PCR directly from 1st-strand or 2nd-strand cDNA using tiled primer pairs in two pools. The entire 29.9 kb viral genome is represented in amplicons of ~400 bp optimized for PE250 sequencing.
Workflow
Lexogen’s SARS-CoV-2 ARTIC panel is a multiplexed amplicon-based whole genome sequencing approach compatible with Illumina sequencing (Figure 2).
Figure 2 | Lexogen ARTIC Amplicon PCR Workflow. The ARTIC panel uses cDNA from SARS-CoV-2 positive samples as input. The sample is split, and amplicon PCR is performed separately for Primer Pool 1 and Primer Pool 2. During the amplicon PCR, Illumina compatible partial sequencing adapters are introduced for a streamlined conversion to sequencing-ready libraries. Following Amplicon PCR, the reaction from Pool 1 and Pool 2 are combined and purified prior to indexing PCR. Up to 384 unique dual indices can be introduced during a 15-minute indexing PCR step, generating ready-to-sequence libraries.
Performance
Primers are based on the two primer sets developed by the ARTIC Network, version N1 (NIID-1)1. The primer sequences are also available on Github as version ver_N1.
Mapping rates vary depended on the concentration and quality of the isolated RNA, the reverse transcription starting from random hexamers, and the high cycle numbers used in the two multiplex PCRs to ensure high amplicon saturation. In a typical example, ~44 % of all reads mapped to the SARS-CoV2 genome (Fig. 3). The coverage reached at least 70x at a read depth of 1.4 Mio mapping reads with a mean coverage of 15,500x.
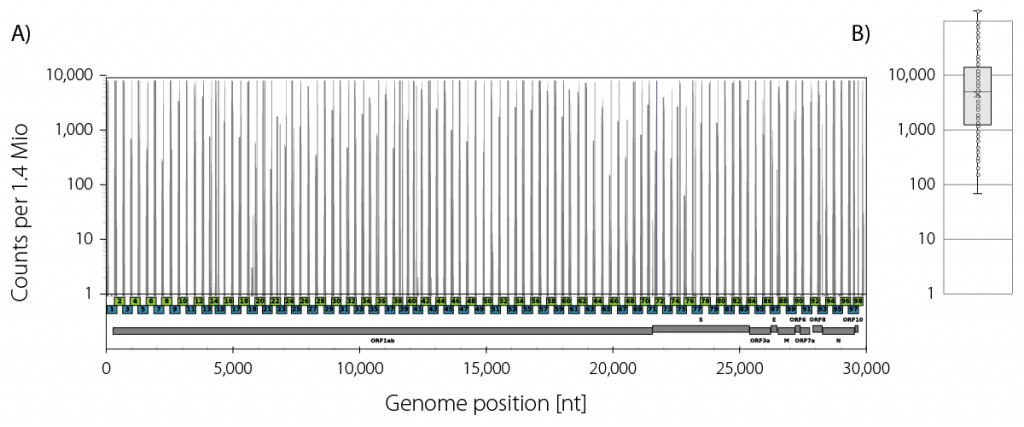
Figure 3 | Lexogen SARS-CoV-2 ARTIC panel coverage profile example. A) Read start site frequencies of the amplicons from both pool 1 (blue) and pool 2 (green) which have an average insert size of 400 bp. B) Box plot of the amplicon frequencies at the given read depth of 1.4 Mio mapping reads with a minimum coverage of 70x.
Specifications
| Feature | Specification |
|---|---|
| Input Material | 1st or 2nd strand cDNA (single- or double-stranded cDNA) |
| Time | ~4.5 h from cDNA to ready-to-sequence library |
| Multiplexing Capacity | Up to 384 Unique Dual Indices |
| Amplicon Size | ~400 bp |
| Sequencing Read Mode | (PE200) – PE250 |
| Recommended Read Depth | min. 1 M reads per sample |
Contact us for more information
For more information on our SARS-CoV-2 products or to request a quote contact Technical Support by using the enquiry form below.



