QuantSeq 3’ mRNA Sequencing Workflow
Step1:
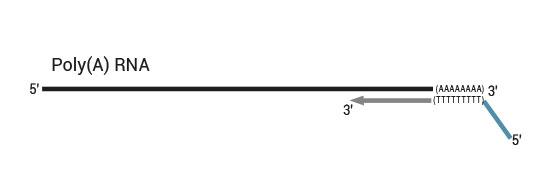
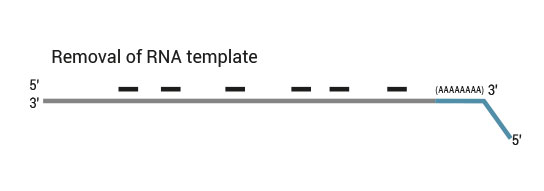
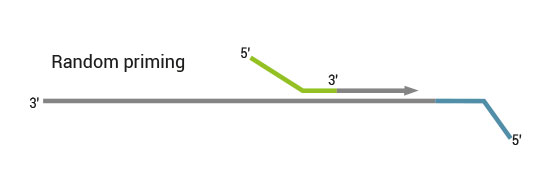
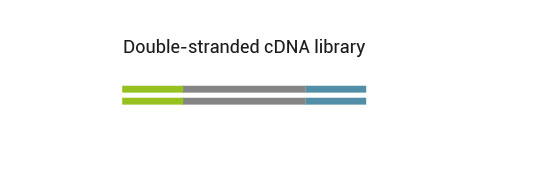
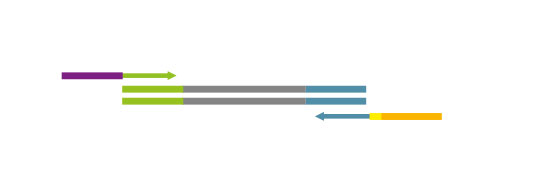
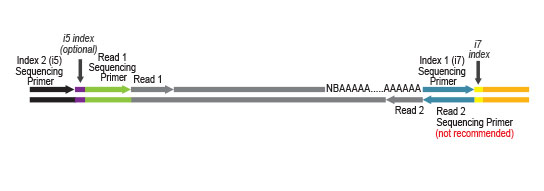
NGS reads are generated towards the poly(A) tail and directly correspond to the mRNA sequence. To pinpoint the exact 3’ end, longer reads may be required (SR100, SR150). Although paired-end sequencing is possible, we do not recommend it for QuantSeq FWD. Read 2 would start with the poly(T) stretch, and as a result of sequencing through the homopolymer stretch, the quality of Read 2 would be very low.