Step 1: Reverse Transcription

![]()
The kit uses total RNA as input, hence no prior poly(A) enrichment or rRNA depletion is needed.
![]()
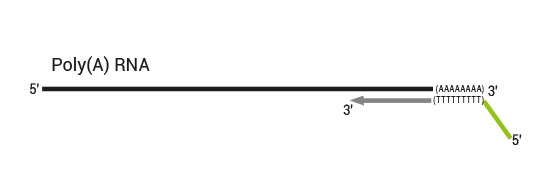
Library generation starts with oligodT priming containing the Illumina-specific Read 1 linker sequence.
Step 2: Removal of RNA
![]()
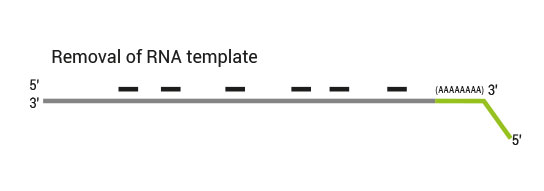
After first strand synthesis the RNA is removed.
Step 3: Second Strand Synthesis
![]()
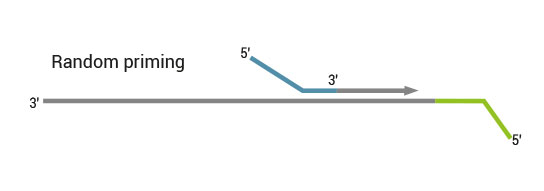
Second strand synthesis is initiated by random priming and a DNA polymerase. The random primer contains the Illumina-specific Read 2 linker sequence.
![]()
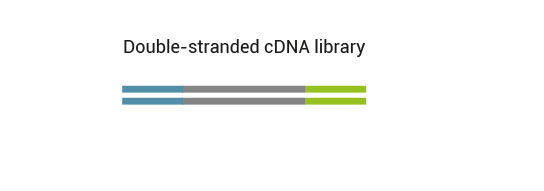
No purification is required between first and second strand synthesis. Second strand synthesis is followed by a magnetic bead-based purification step rendering the protocol compatible with automation.
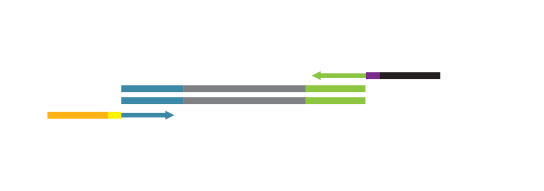
Step 4: Library Amplification
![]()
During the library amplification step sequences required for cluster generation are introduced.
![]()
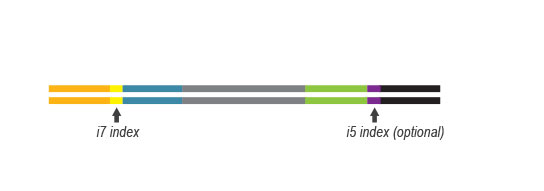
Multiplexing can be performed with up to 384 barcode combinations using the available 96 i7 indices and four i5 indices.
Step 5: Sequencing
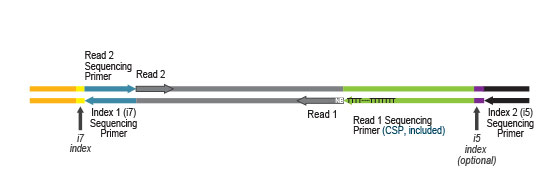
NGS reads are started at the very last nucleotide of the mRNA using the Custom Sequencing Primer (CSP Version 2, included in the kit) for sequencing. The reads generated during Read 1 reflect the cDNA sequence. With QuantSeq REV also paired-end sequencing is possible.



