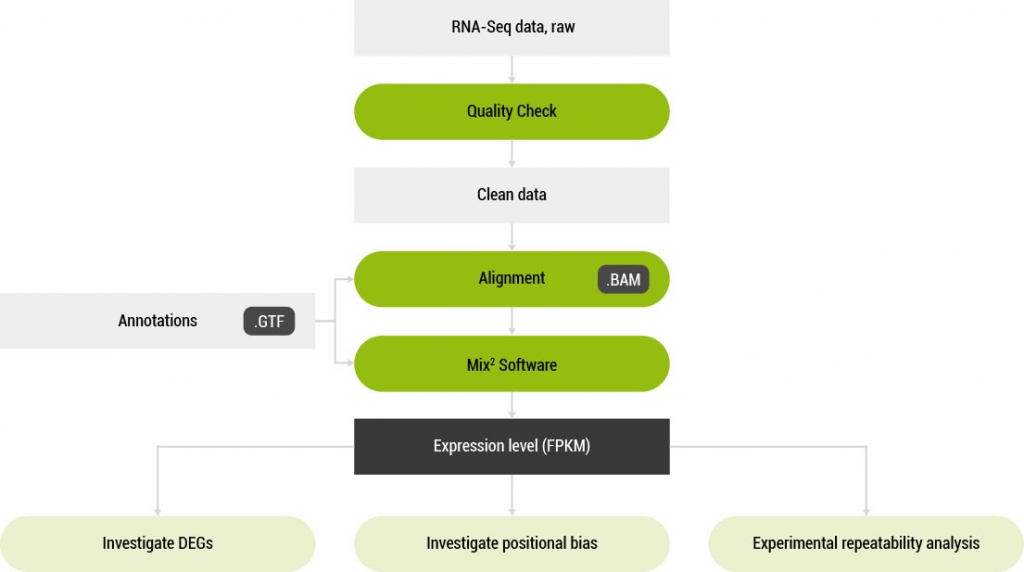
Mix² RNA-Seq Data Analysis Software
- Precise transcript concentration estimates
- Accurate detection of differential expression
- Exceptional reproducibility across variable conditions
- Fast run-times and small memory footprint

Description
Mix² RNA-Seq Data Analysis Software
A software tool for the accurate estimation of RNA concentration from RNA-Seq data.
Model
Fragment bias in RNA-Seq poses a serious challenge to the accurate quantification of gene isoforms. Mix² makes no assumptions about coverage bias but fits for each gene isoform a mixture model to the data (Fig. 1). Mix² can therefore, for instance, accurately represent the 5’ bias, as shown in Fig. 1 (a and b), whereas Cufflinks is restricted to the uniform distribution (Fig. 1c).
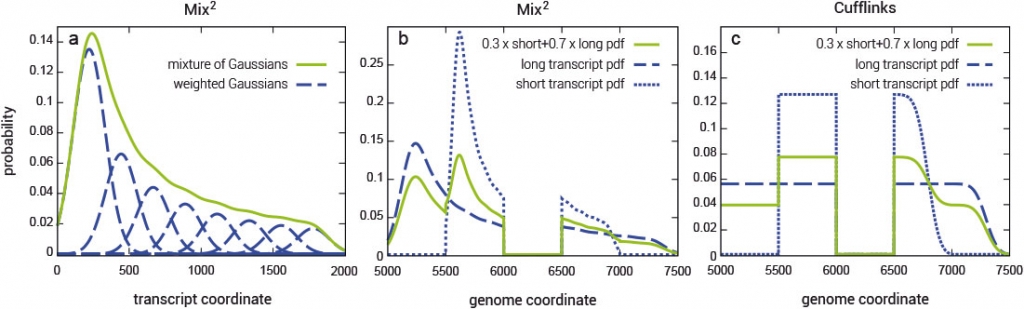
The Mix² software yields accurate isoform quantification from RNA-Seq data
Implementation and run-time performance
The Mix² software runs as a 64-bit Linux command line tool. For an up-to-date list of supported distributions please refer to the User Guide of the Mix² software.
| Mix² | Cufflinks w/o bias correction | Cufflinks with bias correction | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Dataset | Min | GB | Min | xRT | GB | xMEM | Min | xRT | GB | xMEM |
| Avg(UHR) | 7 | 1.26 | 34 | 4.9 | 0.99 | 0.79 | 542 | 77.4 | 1.32 | 1.05 |
| Avg(HBR) | 5 | 1.02 | 32 | 6.4 | 0.90 | 0.88 | 536 | 107.2 | 1.22 | 1.20 |
Table 1 | Memory usage and average run-time statistics on the MAQC UHR and HBR datasets. Min stands for run-time in minutes, GB for memory usage in gigabytes. xRT and xMEM are the factors by which run-time and memory usage increases, respectively, in comparison to Mix².
Featured Publications
Examples
Mix² was tested on the publicly available MicroArray Quality Control (MAQC) [1] and Association of Biomolecular Resource Facilities (ABRF) [2] datasets, containing RNA-Seq data from multiple sequencing facilities and library preparations which started with differently degraded RNA.
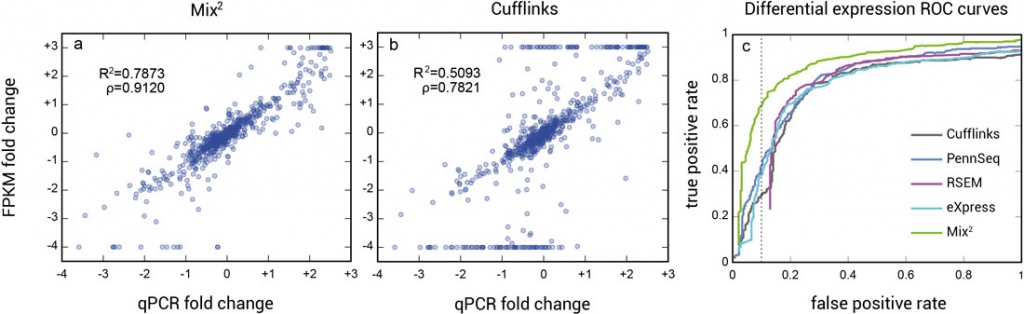
The higher accuracy of the concentration estimates of Mix² leads to better correlation between qPCR and FPKM fold-changes and consequently to higher accuracy in the detection of differential expression (Fig. 2).
FAQ
Frequently Asked Questions
Access our frequently asked question (FAQ) resources via the buttons below.
Please also check our General Guidelines and FAQ resources!
How do you like the new online FAQ resource? Please share your feedback with us!